Je suis Directeur de Recherche INRAE et je m'intéresse à la signalisation nutritionnelle des plantes, en particulier à la régulation du métabolisme azoté et à la voie de signalisation de la kinase TOR (Target of Rapamycin).
Je suis membre de l’équipe NUTS et j’anime depuis 2011 l’Observatoire du Végétal, un réseau labellisé IBiSA de plates-formes technologiques et de phénotypage des plantes (https://www.observatoire-vegetal.inrae.fr/).
I am a member of the NUTS team and since 2011, I have been managing the Plant Observatory, an IBiSA-labeled network of technological and plant phenotyping platforms (https://www.observatoire-vegetal.inrae.fr/) .
Short Biography:
My research career has spanned three decades, starting with a PhD at the Laboratoire de Biologie Cellulaire (INRA Versailles) working on biochemistry and immunochemistry of plant nitrate reductase and other enzymes under the supervision of Pierre Rouzé and Michel Caboche. This was followed in 1988 by a postdoc at the Division of Plant Industry (CSIRO, Canberra) where I worked under the supervision of TJ Higgins on the expression of sulfur-rich proteins in leguminous plants to increase their nutritional value. In 1989, I was recruited as a permanent INRA scientist.
I was deputy Director of the Plant Nitrogen Nutrition Lab (2001-2010) and in 2011 I created the Plant Observatory, a network of plant technological and phenotyping platforms embedded within the Saclay Plant Sciences Lab of Excellence. I first developed PCR-based molecular methods to identify transposons and mutations in nitrate reductase deficient plants. The goal of our group was then to use genetic and molecular tools to identify and characterize components of the nitrate signalling pathway. Twenty years ago, we concentrated on genes in Arabidopsis which were homologous (putative functional orthologs) to genes already known to be regulated by or regulating nitrate assimilation. These candidate genes included NIN-like proteins (NLPs), which are closely related to a transcription factor controlling nodulation in response to nitrate in legume plants. We showed that these transcription factors are master regulators of nitrate assimilation in plants. Another interesting candidate gene was the conserved Target of Rapamycin kinase (TOR), which at this time was already known to be a central regulatory element involved in nitrogen sensing and regulation in yeast and animal cells. Together with the lab of Christophe Robaglia in Cadarache, and later in Marseille, with whom we are still collaborating, we isolated and characterized the first TOR gene in plants. Later, we developed several tools including inducible TOR RNAi lines and phospho-specific antibodies used to measure TOR activity.
The TOR project:
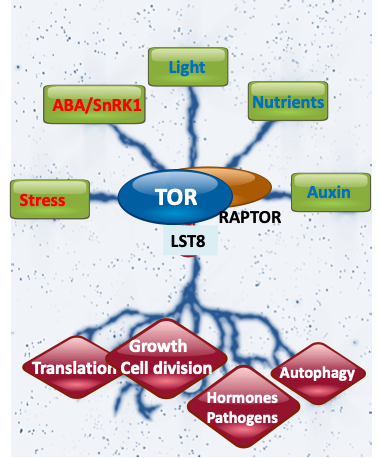
In Eukaryotes, conserved and ancient regulatory circuits allow the essential coupling of cell and tissue growth to the availability of nutrients and energy. TOR is a central regulator of metabolism, mRNA translation, hormone responses and growth processes, promoting growth in favourable conditions while inhibiting catabolism and protein. Plant life depends on a close coupling of many environmental inputs (light and nutrients, but also abiotic or biotic stresses) with complex adaptive growth responses like root and shoot development or the switch from vegetative to reproductive growth. Given the roles of TOR in other Eukaryotes, for example human TOR is linked to cancers or metabolic diseases, this kinase appeared as a good candidate for gaining better knowledge on connections between environmental cues and developmental processes in model and crop plants.
We studied the impact of TOR inactivation on tissue organization and growth and we demonstrated the role of TOR in meristem organization and hypocotyl elongation. Indeed, within the framework of a joint FAPESP-INRA (France-Brazil) project, Chloé Marchive (post-doc) has shown that the activity of TOR is necessary for the elongation of the hypocotyl in the dark ( Zhang et al., 2016 Current Biology) and that the decrease in elongation after TOR inactivation could be reversed by adding brassinosteroids. We also showed that inactivation of TOR resulted in lower accumulation of ABA, proline and raffinose, known to accumulate under stress, and that inactivation of TOR induced enzymes in the catabolism of these metabolites (Kravchenko et al., 2015, BBRC).
As part of a recently completed ANR project (DecoraTOR, 2014-2018), we performed suppressor screens using lst8 mutant to identify new components of the plant TOR signaling pathway. (Forzani et al., 2019 Cell Reports). These screens were supplemented by the identification of mutations conferring resistance to inhibitors of TOR and PI3K kinases (two mutations are under study) and by a double-hybrid screening in yeast using LST8 as bait. So far, six suppressors of the lst8 mutation have been isolated and sequenced in Arabidopsis. Two of these suppressor lines which partially restore growth of lst8 mutants carried mutations in the Yet Another Kinase 1 (AtYAK1) gene encoding a member of the tyrosine / serine dual specific kinase (DYRK) family. Our results revealed that YAK1 is an important TOR kinase effector that transduces the impact of TOR inactivation on growth and metabolism and which probably needs to be turned off to activate plant growth ( Forzani et al., 2019 Cell Reports). YAK1 kinase was first identified in yeast where it is induced by sugar deficiency. Its orthologue in animals is the kinase DYRK1A which is implicated in diseases and mental retardation.
Project:
1- We are developing a project focussed on the roles of transcription factors of the bZIP family on the TOR-dependent transcriptional regulation of genes for the catabolism of proline and raffinose. This project is carried out with Anne-Sophie Leprince (MCU Sorbonne University) and Gustavo Duarte (post-doc until 2019) as part of a project funded by INRA and Agreenskills (ZIPTOR, 2017-2019). For this purpose, Gustavo obtained series of single, double and triple mutants affected in class C and S bZIP factors.
2- We are further characterizing the other lst8 suppressor and AZD8055 resistant/hypersensitive in order to identify new elements of the TOR signaling pathway. We will also pursue the identification of TOR interactors identified by a double hybrid screen by studying their roles in the accumulation of stress-related metabolites through the activity of bZIP transcription factors and in the regulation of growth. We will also study the implications of these signaling pathways in plant branching, which is regulated by nutrient availability and possibly bZIP factors.
3- We are collaborating with Ganna Pansyuk (Necker Enfants Malades, INSERM) on the roles of the conserved class III phosphoinositol-3 kinase (VPS34) in plants and animals. We will further characterize VPS34 inhibitors and mutants to better understand the roles of this kinase in relation with TOR.
Image: Yggdrasil, exploring the nine realms of TOR (from Forzani and Meyer, Nature Plants 2019).
I am a member of the NUTS team and since 2011, I have been managing the Plant Observatory, an IBiSA-labeled network of technological and plant phenotyping platforms (https://www.observatoire-vegetal.inrae.fr/) .
Short Biography:
My research career has spanned three decades, starting with a PhD at the Laboratoire de Biologie Cellulaire (INRA Versailles) working on biochemistry and immunochemistry of plant nitrate reductase and other enzymes under the supervision of Pierre Rouzé and Michel Caboche. This was followed in 1988 by a postdoc at the Division of Plant Industry (CSIRO, Canberra) where I worked under the supervision of TJ Higgins on the expression of sulfur-rich proteins in leguminous plants to increase their nutritional value. In 1989, I was recruited as a permanent INRA scientist.
I was deputy Director of the Plant Nitrogen Nutrition Lab (2001-2010) and in 2011 I created the Plant Observatory, a network of plant technological and phenotyping platforms embedded within the Saclay Plant Sciences Lab of Excellence. I first developed PCR-based molecular methods to identify transposons and mutations in nitrate reductase deficient plants. The goal of our group was then to use genetic and molecular tools to identify and characterize components of the nitrate signalling pathway. Twenty years ago, we concentrated on genes in Arabidopsis which were homologous (putative functional orthologs) to genes already known to be regulated by or regulating nitrate assimilation. These candidate genes included NIN-like proteins (NLPs), which are closely related to a transcription factor controlling nodulation in response to nitrate in legume plants. We showed that these transcription factors are master regulators of nitrate assimilation in plants. Another interesting candidate gene was the conserved Target of Rapamycin kinase (TOR), which at this time was already known to be a central regulatory element involved in nitrogen sensing and regulation in yeast and animal cells. Together with the lab of Christophe Robaglia in Cadarache, and later in Marseille, with whom we are still collaborating, we isolated and characterized the first TOR gene in plants. Later, we developed several tools including inducible TOR RNAi lines and phospho-specific antibodies used to measure TOR activity.
The TOR project:
In Eukaryotes, conserved and ancient regulatory circuits allow the essential coupling of cell and tissue growth to the availability of nutrients and energy. TOR is a central regulator of metabolism, mRNA translation, hormone responses and growth processes, promoting growth in favourable conditions while inhibiting catabolism and protein. Plant life depends on a close coupling of many environmental inputs (light and nutrients, but also abiotic or biotic stresses) with complex adaptive growth responses like root and shoot development or the switch from vegetative to reproductive growth. Given the roles of TOR in other Eukaryotes, for example human TOR is linked to cancers or metabolic diseases, this kinase appeared as a good candidate for gaining better knowledge on connections between environmental cues and developmental processes in model and crop plants.
We studied the impact of TOR inactivation on tissue organization and growth and we demonstrated the role of TOR in meristem organization and hypocotyl elongation. Indeed, within the framework of a joint FAPESP-INRA (France-Brazil) project, Chloé Marchive (post-doc) has shown that the activity of TOR is necessary for the elongation of the hypocotyl in the dark ( Zhang et al., 2016 Current Biology) and that the decrease in elongation after TOR inactivation could be reversed by adding brassinosteroids. We also showed that inactivation of TOR resulted in lower accumulation of ABA, proline and raffinose, known to accumulate under stress, and that inactivation of TOR induced enzymes in the catabolism of these metabolites (Kravchenko et al., 2015, BBRC).
As part of a recently completed ANR project (DecoraTOR, 2014-2018), we performed suppressor screens using lst8 mutant to identify new components of the plant TOR signaling pathway. (Forzani et al., 2019 Cell Reports). These screens were supplemented by the identification of mutations conferring resistance to inhibitors of TOR and PI3K kinases (two mutations are under study) and by a double-hybrid screening in yeast using LST8 as bait. So far, six suppressors of the lst8 mutation have been isolated and sequenced in Arabidopsis. Two of these suppressor lines which partially restore growth of lst8 mutants carried mutations in the Yet Another Kinase 1 (AtYAK1) gene encoding a member of the tyrosine / serine dual specific kinase (DYRK) family. Our results revealed that YAK1 is an important TOR kinase effector that transduces the impact of TOR inactivation on growth and metabolism and which probably needs to be turned off to activate plant growth ( Forzani et al., 2019 Cell Reports). YAK1 kinase was first identified in yeast where it is induced by sugar deficiency. Its orthologue in animals is the kinase DYRK1A which is implicated in diseases and mental retardation.
Project:
1- We are developing a project focussed on the roles of transcription factors of the bZIP family on the TOR-dependent transcriptional regulation of genes for the catabolism of proline and raffinose. This project is carried out with Anne-Sophie Leprince (MCU Sorbonne University) and Gustavo Duarte (post-doc until 2019) as part of a project funded by INRA and Agreenskills (ZIPTOR, 2017-2019). For this purpose, Gustavo obtained series of single, double and triple mutants affected in class C and S bZIP factors.
2- We are further characterizing the other lst8 suppressor and AZD8055 resistant/hypersensitive in order to identify new elements of the TOR signaling pathway. We will also pursue the identification of TOR interactors identified by a double hybrid screen by studying their roles in the accumulation of stress-related metabolites through the activity of bZIP transcription factors and in the regulation of growth. We will also study the implications of these signaling pathways in plant branching, which is regulated by nutrient availability and possibly bZIP factors.
3- We are collaborating with Ganna Pansyuk (Necker Enfants Malades, INSERM) on the roles of the conserved class III phosphoinositol-3 kinase (VPS34) in plants and animals. We will further characterize VPS34 inhibitors and mutants to better understand the roles of this kinase in relation with TOR.
Image: Yggdrasil, exploring the nine realms of TOR (from Forzani and Meyer, Nature Plants 2019).

Contacts
Signalisation, Transport et Utilisation de l'azotehttps://loop.frontiersin.org/people/55357/overview
https://orcid.org/0000-0002-7994-5693
https://www.researchgate.net/profile/Christian-Meyer-18