First systematic analysis in plants of the interaction network formed by recombination initiation proteins: a renewed model
The mechanisms of meiotic CBD formation and the regulations involved are still largely unknown, although it is now clear that the catalytic complex responsible (formed by the proteins SPO11-1, SPO11-2 and MTOPVIB) has strong similarities with certain topoisomerases 1 & 2. In order to understand how the association and activation of the catalytic complex of CBD formation is regulated, we studied the biochemical and genetic interactions existing not only between the CBD-forming proteins, but also between these proteins and other components of the meiotic cellular machinery (chromosome axis, DNA repair proteins etc...).
The results obtained showed that:
> The proteins inducing DNA breaks are organized into several subcomplexes.
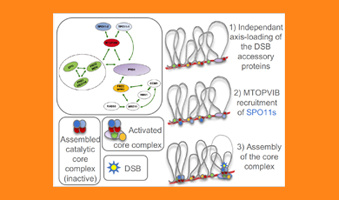
> The MTOPVIB protein occupies a central position in the picture since it is required (i) for the assembly of the catalytic complex 2, (ii) to recruit the catalytic part of the complex to the chromosome axis and (iii) to establish a link between the core proteins and their regulators.
> Among the DSB proteins, PRD3 is likely to play a key regulatory role since it establishes a link between the DSB formation and the DSB repair proteins.
> Finally, our analyses revealed drastic evolutionary divergences for several of the studied proteins.
This work represents a new step towards the understanding of the meiotic recombination initiation mechanisms. It allows to discriminate the catalytic from the regulatory functions involved and to envisage an in vivo and in vitro reconstruction of the DSB formation activity.
The genetic map (sites and rates of crossover formation) is largely determined by the meiotic CBD map. Consequently, the detailed characterization of the mechanisms involved in the initiation of meiotic recombination (nature of the complex, regulation of the assembly of the complex and regulation of its activity) is an essential prerequisite for the development of strategies to modify recombination patterns.
Back
See figure full size below
Legend: The proteins required for CBD formation that initiate meiotic DSB formation are associated in distinct subcomplexes. Most of the CBD-forming proteins are loaded on the chromosome axis independently of each other. Among these, the MTOPVIB protein occupies a central role since (i) it ensures the recruitment to the axis of the catalytic subunits (SPO11-1 and SPO11-2) (ii) it allows the assembly of the catalytic complex and finally (iii) it establishes a connection between the catalytic core subunits and the DSB regulatory proteins.
Scientific Highlight of IJPB
Associated publication:
Vrielynck N, Schneider K, Rodriguez M, JSims J, Chambon A, Hurel A, De Muyt A, Ronceret A, Krsicka O, Mézard C, Schlögelhofer P, Grelon M. Conservation and divergence of meiotic DNA double strand break forming mechanisms in Arabidopsis thaliana. Nucleic Acids Research 2021, 49;7: 9821–9835. doi:0.1093/nar/gkab715