Focus on the APSYNTH team's analytical expertise in a book on the biological degradation of lignins
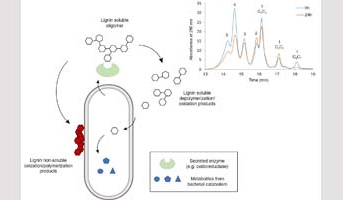
Lignocelluloses are a source of carbon for micro-organisms whose enzymatic systems are able to break down plant cell-wall constituents. Ligninolytic enzymes play a key role in these systems, ensuring cell-wall delignification and increasing cellulose accessibility, an asset for its industrial exploitation.The study of the mechanisms of action of these enzymes on lignin is necessary not only to understand biodegradation, but also to design biocatalysts and biomass transformation pathways in the context of the bioeconomy.
As part of European (Ligbio, Zelcor and Milimo) and ANR (FuncliPro) projects, the "Lignocellulosic Biopolymers: from Cell Wall Assemblies to Synthons for Green Chemistry" APSYNTH team has used its analytical expertise and knowledge of lignin structure and reactivity to study the mechanisms of action of ligninolytic microorganisms and enzymes. A strategy that combines the use of model compounds, the selective extraction of phenolic fractions, their kinetic analysis by steric exclusion chromatography and liquid chromatography-MS, and structural analysis by NMR was developped.
The strategy has enabled the researchers to elucidate the mode of action of bacterial enzymes, with the University of Warwick and CIB-CISC,1-3 fungal enzymes with UMR BBF,4-6 and termite microbiota with UMR IEES.7 The expertise gained through these collaborations has been promoted by a book chapter in a special volume of “Methods in enzymology”.8 It will support the development of future lignocellulosic biorefinery processes.
Références
1Rashid et al. 2018. Sphingobacterium sp. T2 Manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical. ACS Chem. Biol., 13, 2920-2929.
2Rashid et al. 2024. Ether Bond Cleavage of a Phenylcoumaran β-5 Lignin Model Compound and Polymeric Lignin Catalysed by a LigE-type Etherase from Agrobacterium sp. ChemBioChem, 25(8), e202400132.
3Rouches et al. 2021. Assessment strategy for bacterial lignin depolymerization: Kraft lignin and synthetic lignin bioconversion with Pseudomonas putida. Bioresour. Technol. Rep., 15, 100742.
4Daou et al. 2021. A Putative Lignin Copper Oxidase from Trichoderma reesei. J. Fungi, 7, 643.
5Daou et al. 2021. Fungal treatment for the valorization of technical soda lignin. J. Fungi. 7, 39.
6Malric-Garajova et al. 2023. Modification of a Marine Pine Kraft Lignin Sample by Enzymatic Treatment with a Pycnoporus cinnabarinus Laccase. Molecules, 28, 4873.
7Jusselme et al. 2020. Changes in the Phenolic Fraction of Protobind 1000 and Bacterial Microbiota in the Gut of a Higher Termite, Nasutitermes Ephratae. Open access J. Microbiol. Biotechnol.
8Baumberger 2025. Analytical strategies for the investigation of enzymatic and microbial ligninolytic activities. In Methods in Enzymology, Vol. 716 Academic Press, pp. 1-31.
Research developed at the Institute Jean-Pierre Bourgin for Plant Sciences.
Back