Genomic insights into the diversity of cis-regulatory sequences of Arabidopsis thaliana and maize through a comprehensive map of the 5’- and 3’-proximal regions of their genes
transcription
proximal regions of genes
cis-regulatory sequences
computational biology
Arabidopsis thaliana
maize
IJPB featured in a BAP INRAE division highlight, publication of the "Biomass Quality and Interactions with Drought" team
With the development of high-throughput sequencing, the complete sequence of many genomes is now available. Nevertheless, one of the major challenges in genomics is to decode these raw sequences into useful information. In this context, one of the potential levers is the search for cis-regulatory elements, i.e. short DNA sequences that control gene expression1. These sequences are present in high density in the 5’- and 3’-proximal regions of genes surrounding the start and termination sites of transcription, respectively. Characterization of these proximal cis-regulatory sequences is still pending, although experimental and computational work has expanded our knowledge.
With funding from the Plant2Pro® Carnot Institute under the PLMViewer project (2020-2023), we implemented a genome-wide de novo method based on the assumption of topological constraints on the position of cis-regulatory sequences with respect to the transcription start or termination site. Using this approach, a map of preferentially located motifs (PLMs) of Arabidopsis thaliana and maize that reveals the structure and function of proximal cis-regulatory sequences in these two plant species was provided. We report three groups of PLMs in both the 5’- and 3’-proximal regions and emphasize conserved PLMs in both species, particularly in the 3’-proximal region. Comparison with resources from transcription factor and microRNA binding sites shows that 79% of the identified PLMs are unassigned, although some are supported by MNase-defined cistrome occupancy analysis. Enrichment analyses also show that unassigned PLMs provide functional predictions that differ from those derived from transcription factor- and microRNA-binding sites.
Our study illustrates the richness and diversity of PLMs in the proximal regions of Arabidopsis thaliana and maize genes that contribute not only to PLM-transcription factor interactions but also to PLM-microARN and PLM-RNA binding protein interactions2. Detailed analysis of PLMs in other flowering plant species promises to improve the predictive power of breeding models.
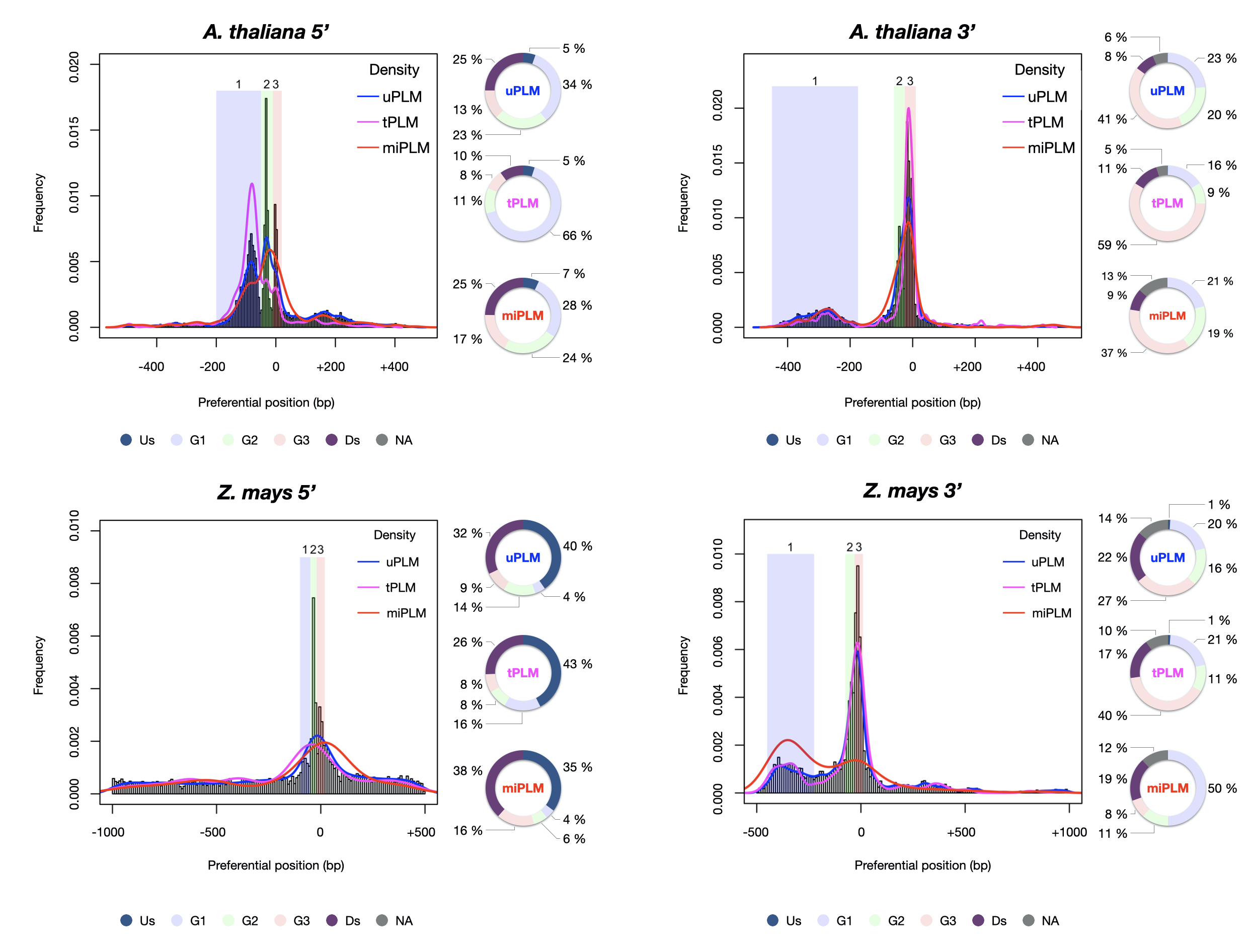
Legend: PLM diversity in 5’- and 3’-proximal regions of Arabidopsis thaliana and maize genes. uPLM : unassigned PLM; tPLM: transcription factor binding sites; miPLM: sequence with homology to microRNA binding sites; Us: region located upstream the three groups; G1, G2 and G3: groups 1, 2 and 3; Ds: region located downstream the three groups; NA: region located between the groups when they are not juxtaposed.
Références
1Schmitz JS, Grotewold E, Stam M (2021). Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. The Plant Cell 34(2): 718–741 https://doi.org/10.1093/plcell/koab281
2Rozière J, Guichard C, Brunaud V, Martin ML, Coursol S (2022). A comprehensive map of preferentially located motifs reveals distinct proximal cis-regulatory sequences in plants. Front. Plant Sci. 12 October 2022 Sec. Plant Bioinformatics https://doi.org/10.3389/fpls.2022.976371
x
With funding from the Plant2Pro® Carnot Institute under the PLMViewer project (2020-2023), we implemented a genome-wide de novo method based on the assumption of topological constraints on the position of cis-regulatory sequences with respect to the transcription start or termination site. Using this approach, a map of preferentially located motifs (PLMs) of Arabidopsis thaliana and maize that reveals the structure and function of proximal cis-regulatory sequences in these two plant species was provided. We report three groups of PLMs in both the 5’- and 3’-proximal regions and emphasize conserved PLMs in both species, particularly in the 3’-proximal region. Comparison with resources from transcription factor and microRNA binding sites shows that 79% of the identified PLMs are unassigned, although some are supported by MNase-defined cistrome occupancy analysis. Enrichment analyses also show that unassigned PLMs provide functional predictions that differ from those derived from transcription factor- and microRNA-binding sites.
Our study illustrates the richness and diversity of PLMs in the proximal regions of Arabidopsis thaliana and maize genes that contribute not only to PLM-transcription factor interactions but also to PLM-microARN and PLM-RNA binding protein interactions2. Detailed analysis of PLMs in other flowering plant species promises to improve the predictive power of breeding models.

Legend: PLM diversity in 5’- and 3’-proximal regions of Arabidopsis thaliana and maize genes. uPLM : unassigned PLM; tPLM: transcription factor binding sites; miPLM: sequence with homology to microRNA binding sites; Us: region located upstream the three groups; G1, G2 and G3: groups 1, 2 and 3; Ds: region located downstream the three groups; NA: region located between the groups when they are not juxtaposed.
Références
1Schmitz JS, Grotewold E, Stam M (2021). Cis-regulatory sequences in plants: Their importance, discovery, and future challenges. The Plant Cell 34(2): 718–741 https://doi.org/10.1093/plcell/koab281
2Rozière J, Guichard C, Brunaud V, Martin ML, Coursol S (2022). A comprehensive map of preferentially located motifs reveals distinct proximal cis-regulatory sequences in plants. Front. Plant Sci. 12 October 2022 Sec. Plant Bioinformatics https://doi.org/10.3389/fpls.2022.976371

x
Back
zoom
IJPB & BAP INRAE division Highlights
"Biomass Quality and Interactions with Drought" team QUALIBIOSEC
Associated publication
Rozière J, Guichard C, Brunaud V, Martin ML, Coursol S (2022).
A comprehensive map of preferentially located motifs reveals distinct proximal cis-regulatory sequences in plants. Front. Plant Sci. 12 October 2022 Sec. Plant Bioinformatics https://doi.org/10.3389/fpls.2022.976371
IJPB & BAP INRAE division Highlights
"Biomass Quality and Interactions with Drought" team QUALIBIOSEC
Associated publication
Rozière J, Guichard C, Brunaud V, Martin ML, Coursol S (2022).
A comprehensive map of preferentially located motifs reveals distinct proximal cis-regulatory sequences in plants. Front. Plant Sci. 12 October 2022 Sec. Plant Bioinformatics https://doi.org/10.3389/fpls.2022.976371