The pentatricopeptide repeat protein MTSF3 is essential for mitochondrial messenger RNA stabilization and embryogenesis in Arabidopsis
mitochondriale respiration
organelle biogenesis
embryo development
Protein PPR
ARNm stability
IJPB featured in a BAP INRAE division highlight, publication of the "Organelles and Reproduction" team
Mitochondria are essential eukaryotic organelles responsible for energy production via aerobic respiration. They are thought to have originated from the endosymbiotic association with an ancient α-proteobacterium. The transition from original endosymbionts to permanent organelles has been accompanied by a massive reduction in their genetic content, a progressive loss of autonomy but limited set of genes essential for mitochondrial activity has been maintained in the organelle. Unlike bacteria, in which gene expression is primarily regulated at the transcriptional level, modern mitochondria possess a surprisingly complex mRNA metabolism involving numerous posttranscriptional RNA modifications that play roles in organellar gene regulation. Gene expression in mitochondria implies a complex cooperation between a degenerated bacterial-like infrastructure (the mitochondrial genome) and nuclear-encoded eukaryotic-derived transfactors, resulting in an RNA metabolism that is far more complex than that of bacteria and which involves many RNA processing steps. These processes rely almost exclusively on nuclear-encoded RNA binding proteins that were acquired during mitonuclear co-evolution. The underlying mechanisms remain however poorly understood at the molecular level and the vast majority of involved protein cofactors have not been identified yet. The most represented type of proteins involved in these processes are the pentatricopeptide repeat (PPR) proteins comprising 2–30 repeats of highly degenerate 31–36 amino acid motifs.
The production of translation-competent mitochondrial mRNAs implies a correct processing of 5’ and 3’ mRNA extremities as well as the stabilization of mature transcripts. The maturation of mRNA ends is particularly essential in plant mitochondria since genes are transcribed from multiple promoters and transcription termination is loosely controlled. Only two PPR proteins involved in the 3’-end processing of the mitochondrial transcripts were reported so far. MITOCHONDRIAL STABILITY FACTOR 1 (MTSF1) and MITOCHONDRIAL STABILITY FACTOR 2 (MTSF2) from Arabidopsis have been shown to be both required for the 3’-end maturation and stability of mature nad4 mRNA and nad1 exon 2-3 precursor RNA, respectively, by associating with the 3’ end of corresponding mitochondrial transcripts.
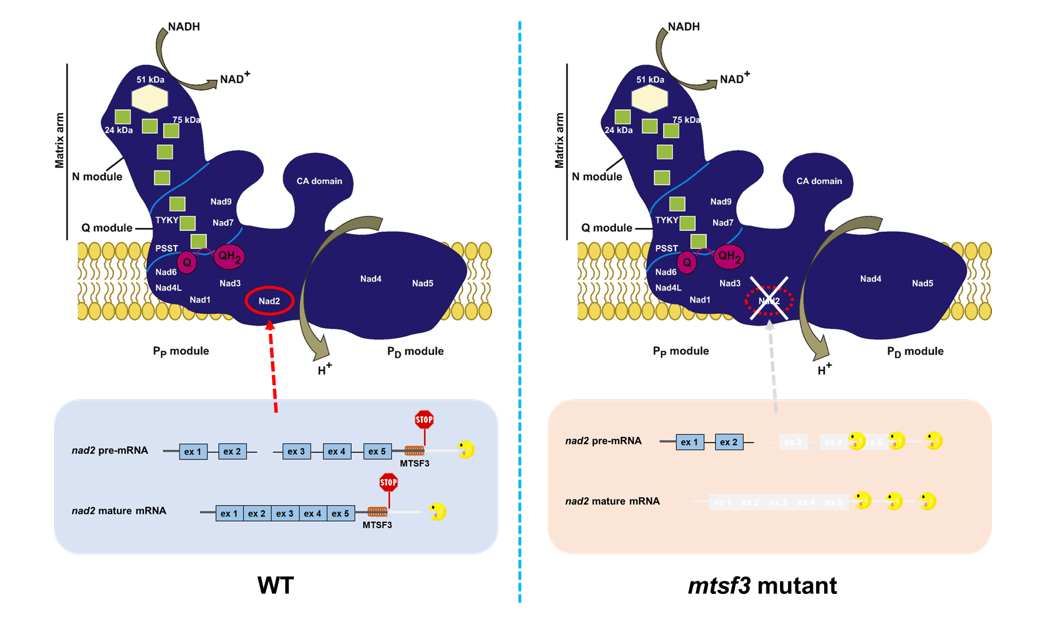
The present study reports on the function of the MITOCHONDRIAL STABILITY FACTOR 3 (MTSF3) protein and shows that it is essential for accumulation of the mitochondrial nad2 transcript (encoding subunit 2 of the mitochondrial NADH dehydrogenase) in Arabidopsis but not for the splicing of nad2 intron 2 as previously proposed. Genetic and biochemical data further demonstrate that the MTSF3 protein associates with high affinity to an RNA region corresponding to the 3’ end of mature nad2 mRNA and that of a nad2 precursor transcript, and exerts a probable protective action by blocking the progression of mitochondrial exoribonucleases. The authors also observed that mtsf3 null mutants were embryo-lethal unlike most Arabidopsis respiratory complex I mutants, suggesting that the generally accepted non-essential character of complex I for embryo development in Arabidopsis needs to be re-evaluated.
Perspectives
MTSF3 protein counts among the very few mRNA stability factors with clear function identified in mitochondria until now. Most identified protein stability factors acting in organelles were shown to stabilize mature messenger RNAs. This study showed that MTSF3 carries two intricately connected functions implying the stabilization of both a pre-mRNA and a mature mRNA. These observations not only advance our knowledge of mRNA 3' end processing in mitochondria, but are also of prime interest for the gene expression community as PPR proteins are present in a wide range of eukaryotes, ranging from yeast to humans.
Références
1Hammani K, Giege P (2014) RNA metabolism in plant mitochondria. Trends Plant Sci 19: 380-389
2Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415-442
3Haili N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H (2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41: 6650-6663
4Wang C, Aube F, Planchard N, Quadrado M, Dargel-Graffin C, Nogue F, Mireau H (2017) The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3’ end of its 5-half intron. Nucleic Acids Res 45: 6119-6134
The production of translation-competent mitochondrial mRNAs implies a correct processing of 5’ and 3’ mRNA extremities as well as the stabilization of mature transcripts. The maturation of mRNA ends is particularly essential in plant mitochondria since genes are transcribed from multiple promoters and transcription termination is loosely controlled. Only two PPR proteins involved in the 3’-end processing of the mitochondrial transcripts were reported so far. MITOCHONDRIAL STABILITY FACTOR 1 (MTSF1) and MITOCHONDRIAL STABILITY FACTOR 2 (MTSF2) from Arabidopsis have been shown to be both required for the 3’-end maturation and stability of mature nad4 mRNA and nad1 exon 2-3 precursor RNA, respectively, by associating with the 3’ end of corresponding mitochondrial transcripts.
The present study reports on the function of the MITOCHONDRIAL STABILITY FACTOR 3 (MTSF3) protein and shows that it is essential for accumulation of the mitochondrial nad2 transcript (encoding subunit 2 of the mitochondrial NADH dehydrogenase) in Arabidopsis but not for the splicing of nad2 intron 2 as previously proposed. Genetic and biochemical data further demonstrate that the MTSF3 protein associates with high affinity to an RNA region corresponding to the 3’ end of mature nad2 mRNA and that of a nad2 precursor transcript, and exerts a probable protective action by blocking the progression of mitochondrial exoribonucleases. The authors also observed that mtsf3 null mutants were embryo-lethal unlike most Arabidopsis respiratory complex I mutants, suggesting that the generally accepted non-essential character of complex I for embryo development in Arabidopsis needs to be re-evaluated.
Perspectives
MTSF3 protein counts among the very few mRNA stability factors with clear function identified in mitochondria until now. Most identified protein stability factors acting in organelles were shown to stabilize mature messenger RNAs. This study showed that MTSF3 carries two intricately connected functions implying the stabilization of both a pre-mRNA and a mature mRNA. These observations not only advance our knowledge of mRNA 3' end processing in mitochondria, but are also of prime interest for the gene expression community as PPR proteins are present in a wide range of eukaryotes, ranging from yeast to humans.
Références
1Hammani K, Giege P (2014) RNA metabolism in plant mitochondria. Trends Plant Sci 19: 380-389
2Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415-442
3Haili N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H (2013) The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res 41: 6650-6663
4Wang C, Aube F, Planchard N, Quadrado M, Dargel-Graffin C, Nogue F, Mireau H (2017) The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3’ end of its 5-half intron. Nucleic Acids Res 45: 6119-6134

Back
Zoom
Legend: Mitochondrial nad2 mRNA is produced as two RNA precursors carrying exons 1-2 and 3-5, respectively. In the presence of the MTSF3 protein, the precursor carrying exons 3-5 and the mature nad2 mRNA are protected from exonuclease progression. In the mstf3 mutant, these two RNAs are degraded.
IJPB & BAP INRAE division Highlights
"Organnelles and Rreproduction" team OrgaRepro
Associated publication
Wang C, Blondel L, Quadrado M, Dargel-Graffin C &Mireau H. (2022). Pentatricopeptide repeat protein MITOCHONDRIAL STABILITY FACTOR 3 ensures mitochondrial RNA stability and embryogenesis. Plant Physiol.190(1): 669-681.
https://doi.org/10.1093/plphys/kiac309