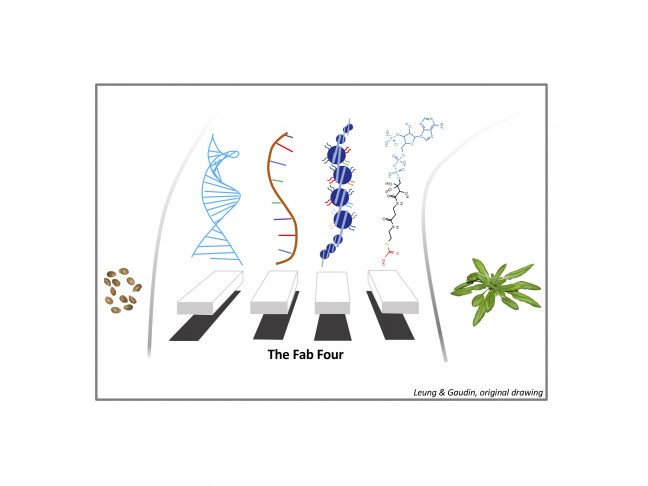
The CHRODYNO group aims to better understand the inter-process coordination between chromatin dynamics and developmental/environmental signaling by exploiting several biological models and multidisciplinary approaches.
Additional Keywords: Arabidopsis, Amaranthus, Chromatin, Epigenetics, Metabolites, Seed, Nucellus, Stress, Drought, evolution
Epigenetics refers to inheritance patterns independent of mutational changes in DNA, which are defined by chemical modifications of the DNA and chromatin-associated proteins. Epigenetic reconfigurations of chromatin determine the diverse transcription outputs from the same genetic blueprint. Epigenetics has thus been argued to be the powerful driving force behind evolution that gave rise to multicellularity, developmental pluripotency and the organism’s capacity to respond to various stresses and adapt to new environmental niches.
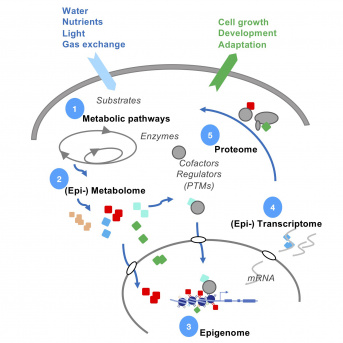
Our major research interests articulate around the interactions between (i) nuclear and chromatin dynamics and (ii) developmental and environmental signaling. We are addressing the question of how epigenetic activities themselves are regulated. One level of coordination must come from their interdependence between the nuclear and the metabolic states of the cell. Indeed, DNA and chromatin proteins are adorned by metabolic moieties, among the most well-known are acetyl and methyl moieties. These same (but not the only) modifications are found also on myriads of proteins, RNA and metabolites, suggesting that they compete with components of epigenetics for the same pool of metabolites that will set the final gene expression patterns [1].
We approach these questions from complementary experimental models by using tools from molecular genetics, biochemistry, cell and structural biology as well as microscopy and modeling.
1. Signalling and seed tissue developmental coordination
Angiosperm seed development is a paradigm of tissue cross-talk as it requires synchronous development of four maternal tissues (nucellus, seed coat, chalaza and funiculus) and two fertilization products (embryo and endosperm).
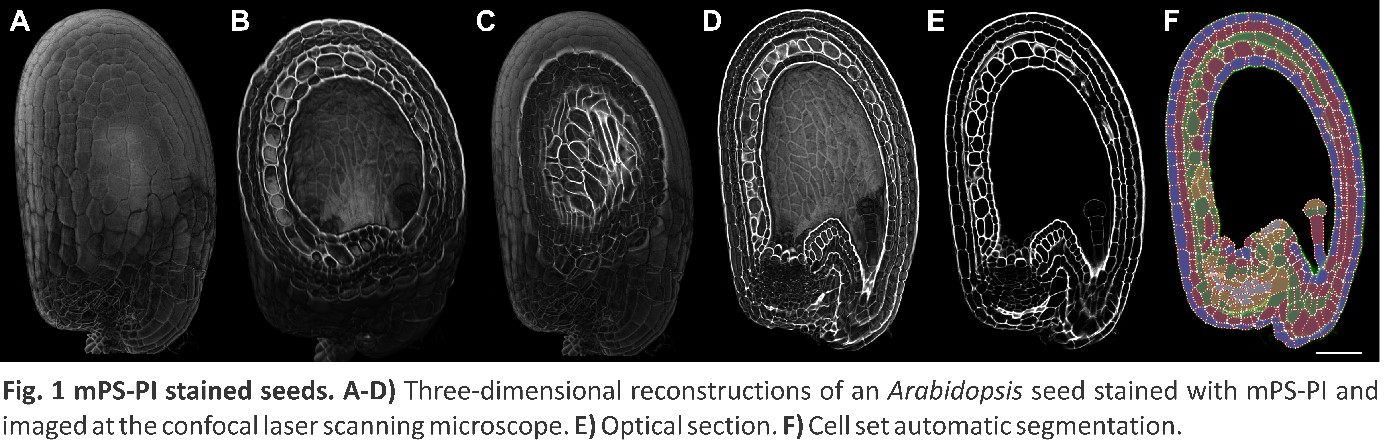
We are characterizing the signaling pathways that coordinate the development of different seed structures and their evolutionary histories, from chromatin organization to downstream signal propagation. We use Arabidopsis and Amaranthus as comparative models for the two most ancient seed structures [2, 3] and exploit an interdisciplinary approach that spans genetics, tissue three-dimensional reconstruction, cell-type-specific transcriptomic profiling, and gene network analysis.
Persons involved Jing Li, PhD student ; Dennys-Marcela Gomez-Paez, post-doctoral fellow ; Valérie Gaudin, scientist ; Enrico Magnani, scientist
2. Coordination of chromatin modification and metabolism in response to environmental stress
We address the functional relationships between metabolism and epigenetics, which are still relatively rarely directly studied in plants.
The current resurgence of excitement on this topic has been revived by the awareness that, in humans, unbalanced diets causing diseases often have their origins in deregulated epigenetic mechanisms, leading to mis-expression of genes. Because many metabolites, histones and chromatin-modifying enzymes are highly conserved, some parallels must exist between plants and humans. On the other hand, because plants have a different lifestyle from that of mammals, how plant-specific biological readouts are shaped, or controlled, by such metabolism-epigenetic interactions will need to be clearly deciphered. In particular, plants produce myriads of metabolites, many of which are implicated in pathogen defense and abiotic stresses, as well as immensely important for human health.
We had identified an epi-metabolite, acetyl 1,3-diaminopropane (acDAP), which is produced by a drought-activated N1-acetyltransferase (NATA1). In guard cells, AcDAP modulates ion transport activities as well as the plasma-membrane potential [4]. The crucial point here is that, in spite of the mild drought, the net physiological output can be stomatal opening, as AcDAP was found to antagonize the action of abscisic acid [5], which triggers stomatal closure to conserve tissue humidity. These results have likely uncovered a mechanism of managing the H2O-CO2 tradeoff to optimize growth when soil-water becomes limited. We are investigating how these diverse plasma-membrane activities controlled by AcDAP are subsequently transmitted to, and coordinated with, the chromatin dynamics.
Persons involved Yann Sulio, undergraduate student ; Elisa Seradin, undergraduate student ; Ekaterina Aleksenko, master student ; Valérie Gaudin, scientist ; Jeffrey Leung, scientist
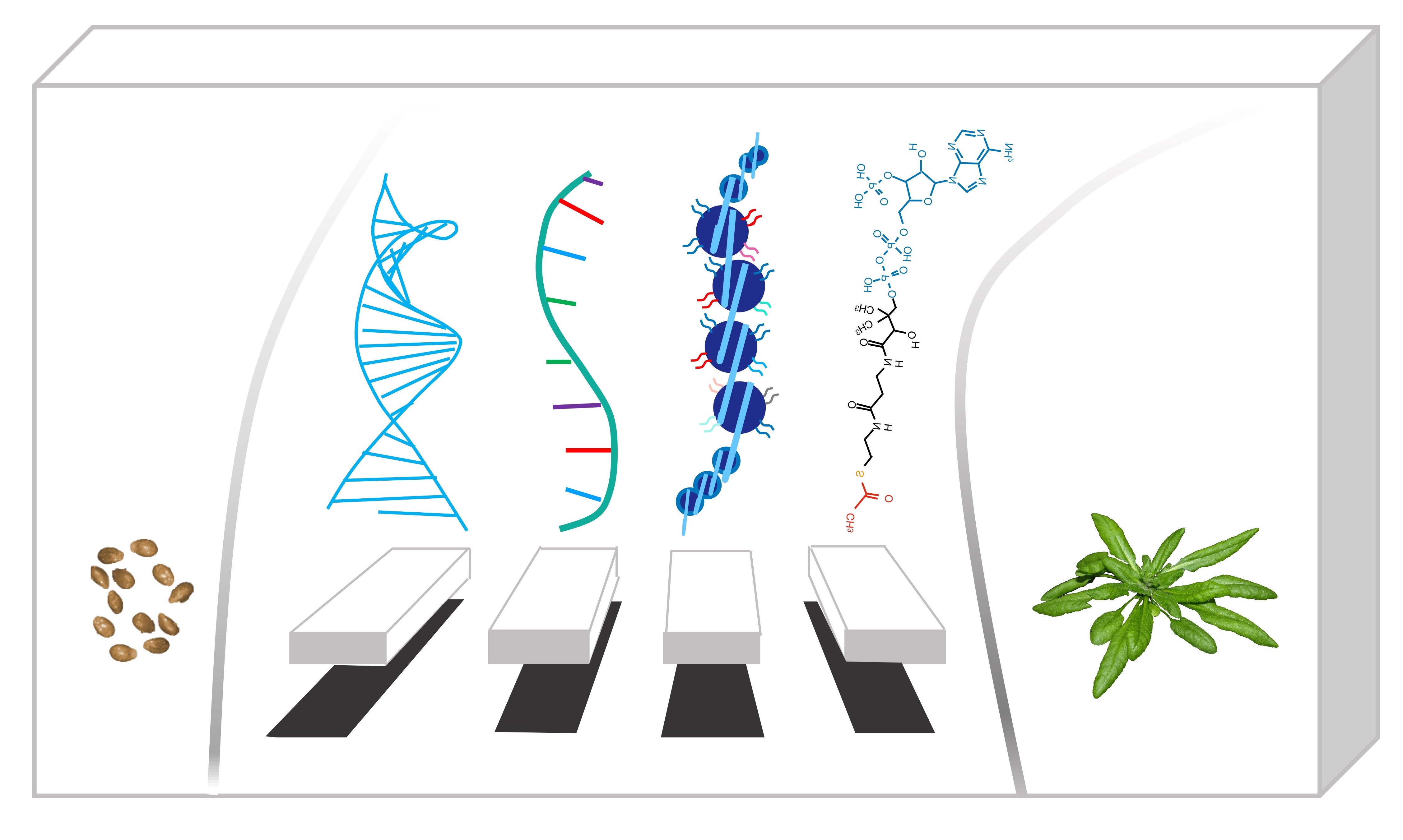
The Fab Four: DNA, RNA, chromatin and metabolites control development and responses to the environment.
3. Modelling nuclear dynamics
Recent progress highlights the amazing complexity, and dynamics as well as structuration of the cell nucleus, in animals and plants, using various tools from microscopy to chromosome conformation capture and biophysics. Whether plants develop specific mechanisms at the scale of the nucleus architecture to allow plant totipotency is an open question. We choose to question the organizational rules of the tri-dimensional nuclear architecture, exploiting the plant model Arabidopsis. We are particularly interested by some nuclear actors, the Polycomb and Trithorax proteins, which play critical dual roles in chromatin dynamics and participate to transcriptional switches. We use an integrative approach combining molecular biology, microscopy and modelling approaches.
Persons involved Enrico Magnani, scientist ; Valérie Gaudin, scientist ; Collaboration Philippe Andrey (IJPB-MIN team)
1. Leung J, Gaudin V: Who Rules the Cell? An Epi-Tale of Histone, DNA, RNA, and the Metabolic Deep State. Front Plant Sci 2020, 11:181.
2. Coen O, Lu J, Xu W, De Vos D, Pechoux C, Domergue F, Grain D, Lepiniec L, Magnani E: Deposition of a cutin apoplastic barrier separating seed maternal and zygotic tissues. BMC Plant Biol 2019, 19(1):304.
3. Magnani E: Seed Evolution, A 'Simpler' Story. Trends Plant Sci 2018, 23(8):654-656.
4. Jammes F, Leonhardt N, Tran D, Bousserouel H, Very AA, Renou JP, Vavasseur A, Kwak JM, Sentenac H, Bouteau F et al: Acetylated 1,3-diaminopropane antagonizes abscisic acid-mediated stomatal closing in Arabidopsis. Plant J 2014, 79(2):322-333.
5. Leung J: The First Broad-Spectrum Abscisic Acid Antagonist. Plant Physiol 2017, 173(4):1939.
6. Molitor A, Latrasse D, Zytnicki M, Andrey P, Houba-Herin N, Hachet M, Battail C, Del Prete S, Alberti A, Quesneville H et al: The Arabidopsis hnRNP-Q Protein LIF2 and the PRC1 subunit LHP1 function in concert to regulate the transcription of stress-responsive genes. Plant Cell 2016, 28:2197-2211.
Epigenetics refers to inheritance patterns independent of mutational changes in DNA, which are defined by chemical modifications of the DNA and chromatin-associated proteins. Epigenetic reconfigurations of chromatin determine the diverse transcription outputs from the same genetic blueprint. Epigenetics has thus been argued to be the powerful driving force behind evolution that gave rise to multicellularity, developmental pluripotency and the organism’s capacity to respond to various stresses and adapt to new environmental niches.
Our major research interests articulate around the interactions between (i) nuclear and chromatin dynamics and (ii) developmental and environmental signaling. We are addressing the question of how epigenetic activities themselves are regulated. One level of coordination must come from their interdependence between the nuclear and the metabolic states of the cell. Indeed, DNA and chromatin proteins are adorned by metabolic moieties, among the most well-known are acetyl and methyl moieties. These same (but not the only) modifications are found also on myriads of proteins, RNA and metabolites, suggesting that they compete with components of epigenetics for the same pool of metabolites that will set the final gene expression patterns [1].
We approach these questions from complementary experimental models by using tools from molecular genetics, biochemistry, cell and structural biology as well as microscopy and modeling.
1. Signalling and seed tissue developmental coordination
Angiosperm seed development is a paradigm of tissue cross-talk as it requires synchronous development of four maternal tissues (nucellus, seed coat, chalaza and funiculus) and two fertilization products (embryo and endosperm).
We are characterizing the signaling pathways that coordinate the development of different seed structures and their evolutionary histories, from chromatin organization to downstream signal propagation. We use Arabidopsis and Amaranthus as comparative models for the two most ancient seed structures [2, 3] and exploit an interdisciplinary approach that spans genetics, tissue three-dimensional reconstruction, cell-type-specific transcriptomic profiling, and gene network analysis.
Persons involved Jing Li, PhD student ; Dennys-Marcela Gomez-Paez, post-doctoral fellow ; Valérie Gaudin, scientist ; Enrico Magnani, scientist

2. Coordination of chromatin modification and metabolism in response to environmental stress
We address the functional relationships between metabolism and epigenetics, which are still relatively rarely directly studied in plants.
The current resurgence of excitement on this topic has been revived by the awareness that, in humans, unbalanced diets causing diseases often have their origins in deregulated epigenetic mechanisms, leading to mis-expression of genes. Because many metabolites, histones and chromatin-modifying enzymes are highly conserved, some parallels must exist between plants and humans. On the other hand, because plants have a different lifestyle from that of mammals, how plant-specific biological readouts are shaped, or controlled, by such metabolism-epigenetic interactions will need to be clearly deciphered. In particular, plants produce myriads of metabolites, many of which are implicated in pathogen defense and abiotic stresses, as well as immensely important for human health.
We had identified an epi-metabolite, acetyl 1,3-diaminopropane (acDAP), which is produced by a drought-activated N1-acetyltransferase (NATA1). In guard cells, AcDAP modulates ion transport activities as well as the plasma-membrane potential [4]. The crucial point here is that, in spite of the mild drought, the net physiological output can be stomatal opening, as AcDAP was found to antagonize the action of abscisic acid [5], which triggers stomatal closure to conserve tissue humidity. These results have likely uncovered a mechanism of managing the H2O-CO2 tradeoff to optimize growth when soil-water becomes limited. We are investigating how these diverse plasma-membrane activities controlled by AcDAP are subsequently transmitted to, and coordinated with, the chromatin dynamics.
Persons involved Yann Sulio, undergraduate student ; Elisa Seradin, undergraduate student ; Ekaterina Aleksenko, master student ; Valérie Gaudin, scientist ; Jeffrey Leung, scientist

The Fab Four: DNA, RNA, chromatin and metabolites control development and responses to the environment.
3. Modelling nuclear dynamics
Recent progress highlights the amazing complexity, and dynamics as well as structuration of the cell nucleus, in animals and plants, using various tools from microscopy to chromosome conformation capture and biophysics. Whether plants develop specific mechanisms at the scale of the nucleus architecture to allow plant totipotency is an open question. We choose to question the organizational rules of the tri-dimensional nuclear architecture, exploiting the plant model Arabidopsis. We are particularly interested by some nuclear actors, the Polycomb and Trithorax proteins, which play critical dual roles in chromatin dynamics and participate to transcriptional switches. We use an integrative approach combining molecular biology, microscopy and modelling approaches.
Persons involved Enrico Magnani, scientist ; Valérie Gaudin, scientist ; Collaboration Philippe Andrey (IJPB-MIN team)

1. Leung J, Gaudin V: Who Rules the Cell? An Epi-Tale of Histone, DNA, RNA, and the Metabolic Deep State. Front Plant Sci 2020, 11:181.
2. Coen O, Lu J, Xu W, De Vos D, Pechoux C, Domergue F, Grain D, Lepiniec L, Magnani E: Deposition of a cutin apoplastic barrier separating seed maternal and zygotic tissues. BMC Plant Biol 2019, 19(1):304.
3. Magnani E: Seed Evolution, A 'Simpler' Story. Trends Plant Sci 2018, 23(8):654-656.
4. Jammes F, Leonhardt N, Tran D, Bousserouel H, Very AA, Renou JP, Vavasseur A, Kwak JM, Sentenac H, Bouteau F et al: Acetylated 1,3-diaminopropane antagonizes abscisic acid-mediated stomatal closing in Arabidopsis. Plant J 2014, 79(2):322-333.
5. Leung J: The First Broad-Spectrum Abscisic Acid Antagonist. Plant Physiol 2017, 173(4):1939.
6. Molitor A, Latrasse D, Zytnicki M, Andrey P, Houba-Herin N, Hachet M, Battail C, Del Prete S, Alberti A, Quesneville H et al: The Arabidopsis hnRNP-Q Protein LIF2 and the PRC1 subunit LHP1 function in concert to regulate the transcription of stress-responsive genes. Plant Cell 2016, 28:2197-2211.
Leader:
Valérie Gaudin