The VAST team (= Variation and Abiotic Stress Tolerance) aims at understanding the genetic bases of adaptation and response to abiotic stress in model plants such as Arabidopsis thaliana.
In that purpose we are essentially using natural variation as a source of biodiversity to discover new genes and new alleles controlling shoot and root development. In other words, our primary interest is in the quantitative genetics of plant growth and genotype x environment interaction (GxE).
Biological Question
The main research lines in the VAST team target a fundamental question in biology: What is the genetic architecture of traits of interest for adaptation and evolution in Arabidopsis ?
We are interested in identifying new factors controlling Arabidopsis growth and fitness in general and their plastic response to the environment in particular, and revealing the molecular variation controlling natural diversity, with no a priori. We benefit from a strong local community involved in studying the biology/physiology behind these interactions, while we tend to focus on a molecular genetics perspective. On the other hand, our experience and knowledge also takes us into the modeling of plant growth responses to the environment.
Additional keywords: Genetic architecture, natural variation, adaptation, transcriptional regulation, growth, plasticity, abiotic stress, combined stress, drought, cold, systems genetics, CRISPR.
Models, tools and methods
For this we use plant growth and its response to the abiotic environment as model complex traits to develop a range of quantitative and molecular genetics approaches using integrative traits as well as traits related to transcriptional and metabolic pathways. Our goal is to understand the molecular and genetic mechanisms underlying variation in plant response to stress caused by drought, nutrient deficiency (N), cold (or their combination), and to fully integrate results at different levels of organisation and scales.
To reach this goal, we combine the technologies, materials and expertise available in the lab (and through collaborations) with novel experimental designs possible only through the use of high-throughput phenotyping and sequencing, bioinformatics and modeling. This involves quantitative genetics approaches such as QTL mapping (using segregating populations, RILs) or GWAS (Genome-Wide Association Studies) to reveal the molecular genetic architecture behind natural variation, but also the exploration of the effect of targeted mutations in different genetic backgrounds (CRISPR...).
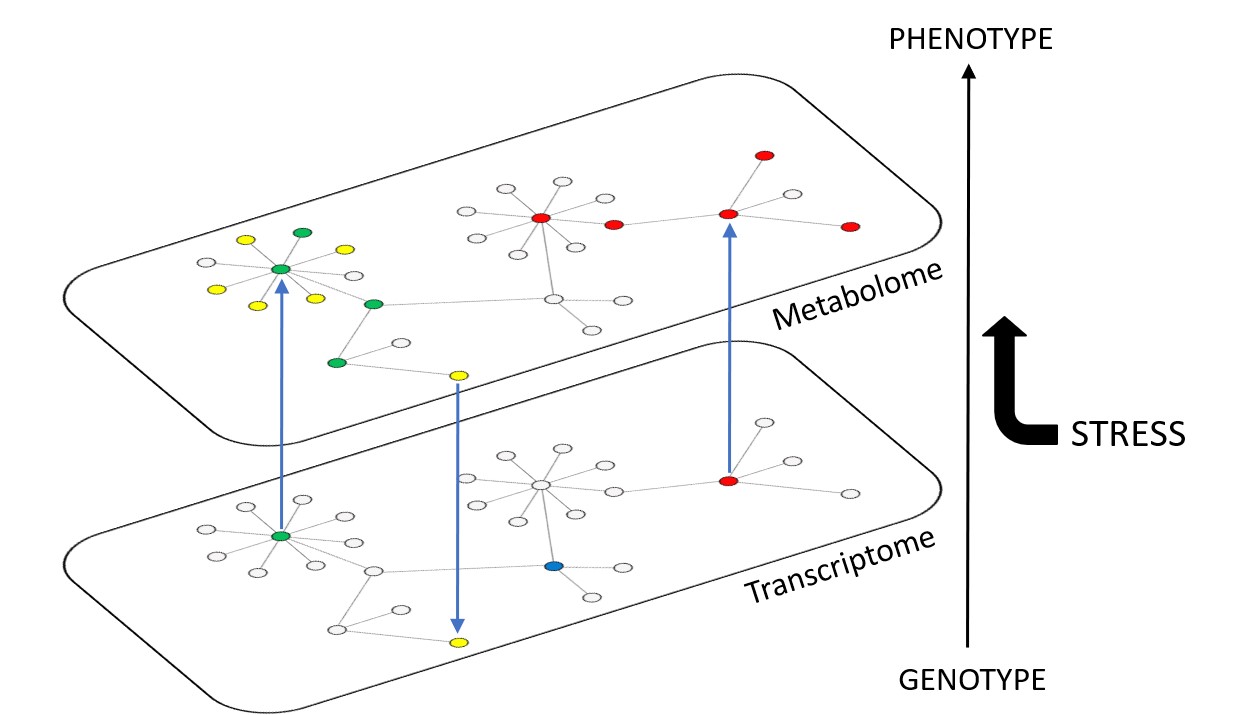
Our model plant of main interest is Arabidopsis thaliana, but we also use relatives such as Cardamine hirsuta. The genetic and physiological complexity revealed here leads us towards mathematical tools to better comprehend the underlying mechanisms at different biological scales (and especially at the metabolomic and proteomic scales). We are refering to 'systems genetics' in our contribution to the development of 'Resource Balance Analysis' metabolic models together with mathematicians, and also in our use of multivariate statistics and network analyses to integrate heterogeneous datasets (phenomics, transcriptomics, metabolomics) and reconstruct causality relationships.
Systems Genetics @ VAST:
Biological Question
The main research lines in the VAST team target a fundamental question in biology: What is the genetic architecture of traits of interest for adaptation and evolution in Arabidopsis ?
We are interested in identifying new factors controlling Arabidopsis growth and fitness in general and their plastic response to the environment in particular, and revealing the molecular variation controlling natural diversity, with no a priori. We benefit from a strong local community involved in studying the biology/physiology behind these interactions, while we tend to focus on a molecular genetics perspective. On the other hand, our experience and knowledge also takes us into the modeling of plant growth responses to the environment.
Additional keywords: Genetic architecture, natural variation, adaptation, transcriptional regulation, growth, plasticity, abiotic stress, combined stress, drought, cold, systems genetics, CRISPR.
Models, tools and methods
For this we use plant growth and its response to the abiotic environment as model complex traits to develop a range of quantitative and molecular genetics approaches using integrative traits as well as traits related to transcriptional and metabolic pathways. Our goal is to understand the molecular and genetic mechanisms underlying variation in plant response to stress caused by drought, nutrient deficiency (N), cold (or their combination), and to fully integrate results at different levels of organisation and scales.
To reach this goal, we combine the technologies, materials and expertise available in the lab (and through collaborations) with novel experimental designs possible only through the use of high-throughput phenotyping and sequencing, bioinformatics and modeling. This involves quantitative genetics approaches such as QTL mapping (using segregating populations, RILs) or GWAS (Genome-Wide Association Studies) to reveal the molecular genetic architecture behind natural variation, but also the exploration of the effect of targeted mutations in different genetic backgrounds (CRISPR...).
Our model plant of main interest is Arabidopsis thaliana, but we also use relatives such as Cardamine hirsuta. The genetic and physiological complexity revealed here leads us towards mathematical tools to better comprehend the underlying mechanisms at different biological scales (and especially at the metabolomic and proteomic scales). We are refering to 'systems genetics' in our contribution to the development of 'Resource Balance Analysis' metabolic models together with mathematicians, and also in our use of multivariate statistics and network analyses to integrate heterogeneous datasets (phenomics, transcriptomics, metabolomics) and reconstruct causality relationships.
Systems Genetics @ VAST:


Leader:
Olivier Loudet













