Cell division in the plant embryo: when a deterministic principle hides behind apparent variability
The positioning of division planes plays a crucial role in shaping the tissular architectures of plants. While many elements involved in plant cell division are well-known, the regulation of division plane positioning remains less understood. The "Modeling and Digital Imaging" MiN team, in collaboration with Jean-Christophe Palauqui "Design, Engineering, Compartmentalization of Lipid Metabolism " DIPOL team and Alain Trubuil from the "Mathematics and Applied Computer Science from Genome to Environment" MaIAGE unit, had previously demonstrated that the observed division planes during the early divisions in the Arabidopsis thaliana model plant embryo corresponded to minimal surfaces passing through the center of the cells1. While division patterns are stereotyped up to the 16-cell stage, they become variable beyond that point. The question then arose to characterize this variability and determine if it corresponded to a new division rule.
Combining 3D microscopy and an original quantitative image analysis approach, the researchers showed that the variability in the orientation of division planes in the Arabidopsis thaliana embryo is paradoxically accompanied by a homogenization of cell shapes. Computer modeling demonstrated that most variable divisions observed beyond the 16-cell stage could be predicted by the same deterministic rule as the initial stereotyped divisions. The researchers also highlighted the introduction, at the 16-cell stage, of cell geometries quasi-invariant under rotation. Hence, by linking cell geometry to the positioning of the division plane, the modeling solved the paradox between variability in the orientation of cell divisions and homogenization of cell shapes. The results indicate that tissue development in the Arabidopsis thaliana embryo follows a self-organization principle connecting cell shape and division orientation, suggesting a remarkable economy of means in controlling embryonic division orientation.
This research underscores the importance of self-organization processes in plant embryonic development and illustrates how modeling contributes to deciphering the apparent complexity of biological systems. The developed approaches will help elucidate the principles underlying tissue development in different systems and experimental conditions.
1 J Moukhtar, A Trubuil, K Belcram, D Legland, Z Khadir, A Urbain, J-C Palauqui & P Andrey (2019). Cell geometry determines symmetric and asymmetric division plane selection in Arabidopsis early embryos. PLoS Computational Biology, 15, e1006771.
https://doi.org/10.1371/journal.pcbi.1006771
Back
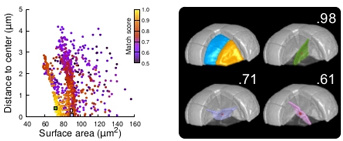
Legend: Modeling of division patterns (here at 5th division generation) in inner cells based on geometric features. Left: distribution of simulation results in an internal apical cell (N=1000). Simulated planes are characterized by their area and distance from the geometric center of the mother cell. Color indicates the degree of matching between simulated and observed planes. Right: observed daughter cells (blue and orange); three simulated planes are shown in the reconstructed mother cell (Transparent). Green: simulation best matching the observed plane. Purple and pink: simulations with alternative orientations. Numbers indicate degrees of matching. Red dot: center of mother cell. In the graph on the left, the positions of the three simulated planes are represented by squares of the same color.
IJPB & BAP INRAE division Highlight
"Modeling and Digital Imaging" MiN team
Associated publication
E Laruelle, K Belcram, A Trubuil, JC Palauqui and P Andrey (2022). Large-scale analysis and computer modeling reveal hidden regularities behind variability of cell division patterns in Arabidopsis thaliana embryogenesis. Elife, 11, e79224.
https://doi.org/10.7554/eLife.79224