Our research aims at designing original approaches in image analysis, spatial statistics, and computational modeling to better understand plant development and functions.
*** Post-doc position currently available, another to open soon (see Jobs) ***
Motivation
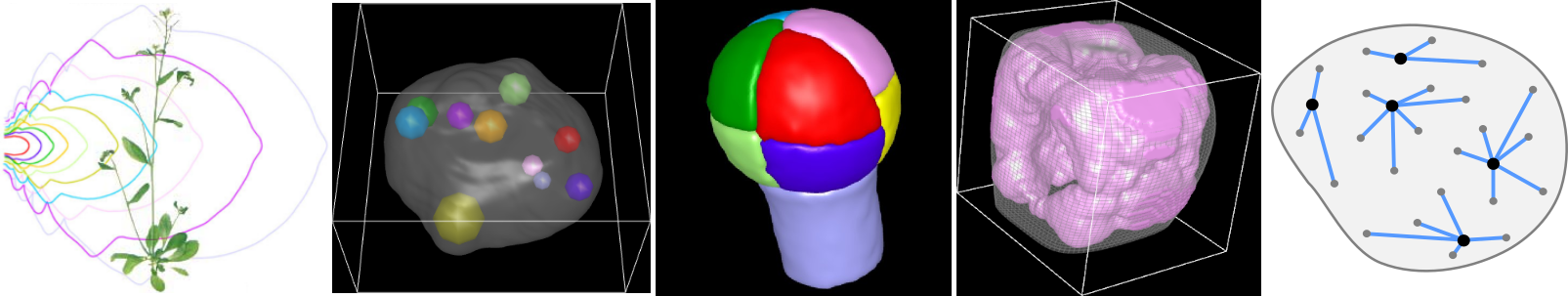
A major question in plant developmental biology is to understand how various factors, processes and scales are integrated during morphogenesis to build specific organization patterns and shapes at the levels of tissues and of organs. We address this questionning in our research activities by developing new methods for visualizing, quantifying, and modeling the data provided by microscopy images of plant systems.
Themes
Our research activities revolve around three main axes:
- image processing and analysis: we develop algorithmic strategies to extract quantitative information from microscopy images and to build statistical representations from series of individual observations;
- spatial statistics: we develop methods to statistically map in 3D spatial organizations in biological systems and to dissect the architectural principles that subtend these organizations;
- computational modeling: we develop predictive computational models to understand the mechanisms that are involved in the establishment and in the regulation of spatial patterns and of plant organ shapes.
Motivation
A major question in plant developmental biology is to understand how various factors, processes and scales are integrated during morphogenesis to build specific organization patterns and shapes at the levels of tissues and of organs. We address this questionning in our research activities by developing new methods for visualizing, quantifying, and modeling the data provided by microscopy images of plant systems.
Themes
Our research activities revolve around three main axes:
- image processing and analysis: we develop algorithmic strategies to extract quantitative information from microscopy images and to build statistical representations from series of individual observations;
- spatial statistics: we develop methods to statistically map in 3D spatial organizations in biological systems and to dissect the architectural principles that subtend these organizations;
- computational modeling: we develop predictive computational models to understand the mechanisms that are involved in the establishment and in the regulation of spatial patterns and of plant organ shapes.

Selected publications (see 'Publications' tab for complete list)
Laruelle E, Belcram K, Trubuil A, Palauqui J-C and Andrey P (2022). Large-scale analysis and computer modeling reveal hidden regularities behind variability of cell division patterns in Arabidopsis thaliana embryogenesis. eLife, 11, e79224. https://doi.org/10.7554/eLife.79224.
Arpòn J, Sakai K, Gaudin V and Andrey P (2021). Spatial modeling of biological patterns shows multiscale organization of Arabidopsis thaliana heterochromatin. Scientific Reports, 11, 323. https://doi.org/10.1038/s41598-020-79158-5.
Burguet J and Andrey P (2020). Edge correction for intensity estimation of 3D heterogeneous point processes from replicated data. Spatial Statistics, 36, 100421. https://doi.org/10.1016/j.spasta.2020.100421.
Moukhtar J, Trubuil A, Belcram K, Legland D, Khadir Z, Urbain A, Palauqui J-C and Andrey P (2019). Cell geometry determines symmetric and asymmetric division plane selection in Arabidopsis early embryos. PLoS Computational Biology, 15, e1006771. https://doi.org/10.1371/journal.pcbi.1006771.
Arganda-Carreras I and Andrey P (2017). Designing Image Analysis Pipelines in Light Microscopy: A Rational Approach. Methods in Molecular Biology, 1563, 185-207. https://doi.org/10.1007/978-1-4939-6810-7_13.
Biot E, Cortizo M, Burguet J, Kiss A, Oughou M, Maugarny-Calès A, Gonçalves B, Adroher B, Andrey P, Boudaoud A and Laufs P (2016). Multiscale quantification of morphodynamics: MorphoLeaf, software for 2-D shape analysis. Development, 143, 3417-3428. https://doi.org/10.1242/dev.134619.
Biot E, Crowell E, Burguet J, Höfte H, Vernhettes S and Andrey P (2016). Strategy and software for the statistical spatial analysis of 3D intracellular distributions. Plant Journal, 87, 230-242. https://doi.org/10.1111/tpj.13189.
Legland D, Arganda-Carreras I and Andrey P (2016). MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics, 32, 3532-3534. https://doi.org/10.1093/bioinformatics/btw413.
Burguet J and Andrey P (2014). Statistical comparison of spatial point patterns in biological imaging. PLoS ONE, 9, e87759. https://doi.org/10.1371/journal.pone.0087759.
Andrey P, Kiêu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot P-G, Hue-Beauvais C, Houba-Hérin N, Duranthon V, Devinoy E, Beaujean N, Gaudin V, Maurin Y and Debey P (2010). Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Computational Biology, 6, e1000853. https://doi.org/10.1371/journal.pcbi.1000853.
Andrey P and Maurin Y (2005). Free-D: an integrated environment for three-dimensional reconstruction from serial sections. Journal of Neuroscience Methods, 145, 233-244. https://doi.org/10.1016/j.jneumeth.2005.01.006.
Laruelle E, Belcram K, Trubuil A, Palauqui J-C and Andrey P (2022). Large-scale analysis and computer modeling reveal hidden regularities behind variability of cell division patterns in Arabidopsis thaliana embryogenesis. eLife, 11, e79224. https://doi.org/10.7554/eLife.79224.
Arpòn J, Sakai K, Gaudin V and Andrey P (2021). Spatial modeling of biological patterns shows multiscale organization of Arabidopsis thaliana heterochromatin. Scientific Reports, 11, 323. https://doi.org/10.1038/s41598-020-79158-5.
Burguet J and Andrey P (2020). Edge correction for intensity estimation of 3D heterogeneous point processes from replicated data. Spatial Statistics, 36, 100421. https://doi.org/10.1016/j.spasta.2020.100421.
Moukhtar J, Trubuil A, Belcram K, Legland D, Khadir Z, Urbain A, Palauqui J-C and Andrey P (2019). Cell geometry determines symmetric and asymmetric division plane selection in Arabidopsis early embryos. PLoS Computational Biology, 15, e1006771. https://doi.org/10.1371/journal.pcbi.1006771.
Arganda-Carreras I and Andrey P (2017). Designing Image Analysis Pipelines in Light Microscopy: A Rational Approach. Methods in Molecular Biology, 1563, 185-207. https://doi.org/10.1007/978-1-4939-6810-7_13.
Biot E, Cortizo M, Burguet J, Kiss A, Oughou M, Maugarny-Calès A, Gonçalves B, Adroher B, Andrey P, Boudaoud A and Laufs P (2016). Multiscale quantification of morphodynamics: MorphoLeaf, software for 2-D shape analysis. Development, 143, 3417-3428. https://doi.org/10.1242/dev.134619.
Biot E, Crowell E, Burguet J, Höfte H, Vernhettes S and Andrey P (2016). Strategy and software for the statistical spatial analysis of 3D intracellular distributions. Plant Journal, 87, 230-242. https://doi.org/10.1111/tpj.13189.
Legland D, Arganda-Carreras I and Andrey P (2016). MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics, 32, 3532-3534. https://doi.org/10.1093/bioinformatics/btw413.
Burguet J and Andrey P (2014). Statistical comparison of spatial point patterns in biological imaging. PLoS ONE, 9, e87759. https://doi.org/10.1371/journal.pone.0087759.
Andrey P, Kiêu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot P-G, Hue-Beauvais C, Houba-Hérin N, Duranthon V, Devinoy E, Beaujean N, Gaudin V, Maurin Y and Debey P (2010). Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Computational Biology, 6, e1000853. https://doi.org/10.1371/journal.pcbi.1000853.
Andrey P and Maurin Y (2005). Free-D: an integrated environment for three-dimensional reconstruction from serial sections. Journal of Neuroscience Methods, 145, 233-244. https://doi.org/10.1016/j.jneumeth.2005.01.006.
Leader:
Philippe Andrey