Contrasting epigenetic regulation of transgenes and endogenous genes in the plant Arabidopsis thaliana
transgene
RNA silencing
siRNA
DNA methylation
histone
chromatin
In the context of predictive biology approaches and scientific expertise for to inform public policies, it is essential to understand the role of the different chromatin epigenetic marks.
The transfer of genetic material by crossing or transgenesis, as well as the modification of genetic information by mutagenesis, can lead to local or global modifications of the genomice's epigenetic marks. It is therefore essential to understand the impact of these epigenetic modifications and precise the role of each of these marks. The epigenetic impact of genome modifications obtained by CRISPR technology must also need to be investigated.
For almost 40 years, the behavior of transgenic plants has been studied extensively. Depending on where they integrate in the genome, transgenes can be recognized as intrusive DNA sequences and silenced, or can be expressed stably over generations.
Transgenes that are stably expressed in plants over several generations are generally considered as assimilated by the genome and epigenetically similar to endogenous genes. In this study, Hervé Vaucheret's team, EpiARN in collaboration with Bernard J. Carroll, The University of Queensland, Brisbane, Australia, shows that IBM1, an enzyme that demethylates lysine 9 of histone H31 does not counteract DNA methylation of endogenous genes but not transgenes. It is JMJ14, an enzyme that demethylates lysine 4 of histone H3 without affection DNA methylation of endogenous genes, which limits transgene DNA methylation 2. These results therefore indicate that transgenes undergo epigenetic regulations that differ from those from endogenous genes
Attenuation of transgene DNA methylation by JMJ14 promotes the production of aberrant RNAs that can induce transgene posttranscriptional silencing. These results therefore indicate that, through JMJ14, transgenes remain susceptible to silencing, even when stably expressed. The maintenance of stably expressed transgenes in a probationary state of silencing susceptibility suggests that plants impose epigenetic marks on transgenes to durably distinguish them from endogenous genes.
Outlook
This study was conducted on transgenes consisting exclusively of exogenous sequences. To determine if it is their exogenous nature or the local disruption of genome organization occurring during integration that makes transgenes epigenetically different from endogenous genes, similar experiments are now planed with transgenes consisting of endogenous sequences. The epigenetic behavior of endogenous sequences modified by the CRISPR technology will also be studied, as well as that of new copies of transposable elements.
Références
1 Miura, A., Nakamura, M., Inagaki, S., Kobayashi, A., Saze, H., and Kakutani, T. (2009). An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28, 1078-1086.
2 Le Masson, I., Jauvion, V., Bouteiller, N., Rivard, M., Elmayan, T., and Vaucheret, H. (2012). Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24, 3603-361
For almost 40 years, the behavior of transgenic plants has been studied extensively. Depending on where they integrate in the genome, transgenes can be recognized as intrusive DNA sequences and silenced, or can be expressed stably over generations.
Transgenes that are stably expressed in plants over several generations are generally considered as assimilated by the genome and epigenetically similar to endogenous genes. In this study, Hervé Vaucheret's team, EpiARN in collaboration with Bernard J. Carroll, The University of Queensland, Brisbane, Australia, shows that IBM1, an enzyme that demethylates lysine 9 of histone H31 does not counteract DNA methylation of endogenous genes but not transgenes. It is JMJ14, an enzyme that demethylates lysine 4 of histone H3 without affection DNA methylation of endogenous genes, which limits transgene DNA methylation 2. These results therefore indicate that transgenes undergo epigenetic regulations that differ from those from endogenous genes
Attenuation of transgene DNA methylation by JMJ14 promotes the production of aberrant RNAs that can induce transgene posttranscriptional silencing. These results therefore indicate that, through JMJ14, transgenes remain susceptible to silencing, even when stably expressed. The maintenance of stably expressed transgenes in a probationary state of silencing susceptibility suggests that plants impose epigenetic marks on transgenes to durably distinguish them from endogenous genes.
Outlook
This study was conducted on transgenes consisting exclusively of exogenous sequences. To determine if it is their exogenous nature or the local disruption of genome organization occurring during integration that makes transgenes epigenetically different from endogenous genes, similar experiments are now planed with transgenes consisting of endogenous sequences. The epigenetic behavior of endogenous sequences modified by the CRISPR technology will also be studied, as well as that of new copies of transposable elements.
Références
1 Miura, A., Nakamura, M., Inagaki, S., Kobayashi, A., Saze, H., and Kakutani, T. (2009). An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28, 1078-1086.
2 Le Masson, I., Jauvion, V., Bouteiller, N., Rivard, M., Elmayan, T., and Vaucheret, H. (2012). Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 24, 3603-361
Back
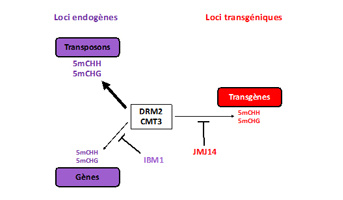
Legend: DRM2 and CMT3 methylate CHG and CHH sites in plant DNA and silence transposons. IBM1 (an histone demethylase acting on lysine 9 of histone H3) counteracts endogenous gene methylation, while JMJ14 JMJ14 (an histone demethylase acting on lysine 4 of histone H3) counteracts transgene methylation and promotes their post-transcriptional inactivation.
Scientific Highlight of IJPB
"Epigenetics and Small RNAs" team EpiARN
Associated publication
Butel, N., Yu, A., Le Masson, I., Borges, F., Elmayan, T., Taochy, C., Gursanscky, N.R., Cao, J., Bi, S., Sawyer, A., Carroll, B.J., and Vaucheret, H. (2021). Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis. Nat Commun 12, 2787. doi : 10.1038/s41467-021-22995-3
Scientific Highlight of IJPB
"Epigenetics and Small RNAs" team EpiARN
Associated publication
Butel, N., Yu, A., Le Masson, I., Borges, F., Elmayan, T., Taochy, C., Gursanscky, N.R., Cao, J., Bi, S., Sawyer, A., Carroll, B.J., and Vaucheret, H. (2021). Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis. Nat Commun 12, 2787. doi : 10.1038/s41467-021-22995-3