Essential Role of the “Shuttle” Protein SGS3 in Gene Silencing in the Plant Arabidopsis thaliana
A genetic screen performed in planta identified the key players involved in the cytoplasmic steps of RNAi: the RDR6 protein, which converts aberrant RNAs into double-stranded RNA; DCL proteins, which cleave double-stranded RNA into siRNAs; and the AGO1 protein, which loads siRNAs to guide the degradation of matching mRNAs (see figure). However, the mechanism by which aberrant RNAs are selected in the nucleus and transported to the RNAi machinery in the cytoplasm remained unclear. The article published in Nature Communications shows that the SGS3 protein shuttles between the nucleus and the cytoplasm. In the nucleus, SGS3 interacts with the chromatin remodeling factor CHR11 and binds to aberrant RNAs. In the cytoplasm, it interacts with the RNA polymerase RDR6. Sequencing of small RNAs produced when the RNA quality control machinery is disrupted by a viral infection revealed that the vast majority of genes generating these small RNAs recruit CHR11. This study led to a model in which SGS3 binds to aberrant RNAs transcribed in the nucleus from genes associated with CHR11, and exports them to the cytoplasm, where they are converted into double-stranded RNA by RDR6 to initiate the production of siRNAs—either across a large portion of the genome during viral infection, or in a gene-specific manner following the introduction of a transgene.
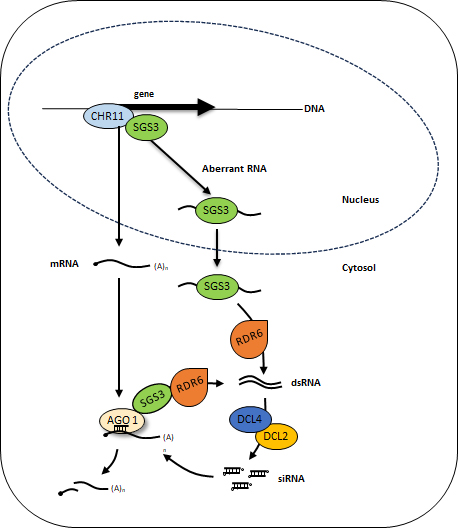
Identifying endogenous genes that are likely to engage in the RNAi pathway when the plant's RNA quality control system is disrupted helps to flag potential silencing risks. These risks can arise from changes in the expression of such genes through transgenesis, or even through targeted genome editing.
Research developed at the Institute Jean-Pierre Bourgin for Plant Sciences in collaboration.
Back
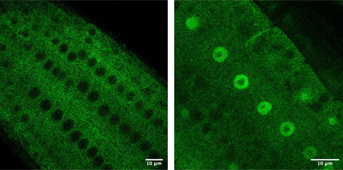
Légend:
Left: diffuse localization of SG3-GFP (marked in green) in thale cress root cells
Right: blockage of SGS3-GFP (marked in green) in the nucleus of thale cress root cells (after treatment with a nuclear export inhibitor)
IJPB Highlight
Référence:
Elmayan T, Blein, T, Elvira-Matelot, E., Le Masson, I, Christ, A, Bouteiller, N, Crespi, MD, Vaucheret, H. Arabidopsis SGS3 is recruited to chromatin by CHR11 to select RNA that initiate siRNA production. Nature Communications 2025.
doi : https://doi.org/10.1038/s41467-025-57394-5
Contact: Hervé Vaucheret, contact
IJPB team:
"Epigenetics and small RNAs" epiARN team
IPS2 collaborating team:
"Regulatory non-coding RNAs in root plasticity" REGARN