SAS team from IJPB paves the way for the comprehension of allelopathic interactions
Faced with the challenge of the agro-ecological transition and the massive reduction in the use of pesticides, exploiting the 'allelopathic' properties of crops appears to be a judicious approach towards the biological regulation of weeds. Allelopathy is a mechanism by which a plant, via the release of specialized metabolites, positively or negatively affects the growth of neighboring plants independently of trophic relationships. Nonetheless, promoting weed control through the mobilization of allelopathic properties remains a complex process to implement. Indeed, the effects of allelopathy (per se) are difficult to qualify and quantify on target plants due to the complexity of plant interactions such as competition for resources. Moreover, neither the molecular mechanisms involved, nor the genetic and environmental control of allelopathic effects are well characterised.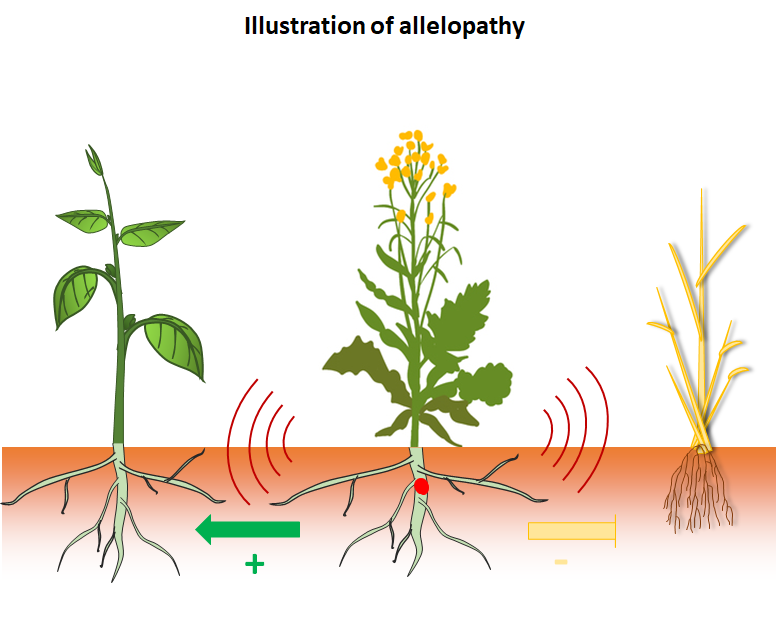
The SAS (Allelopathic Signaling and Strigolactone) team at the IJPB aims to identify the genes and chemical signals in the rhizosphere involved in plant-plant interactions and to understand how these signals can affect the development of other plants in order to contribute to the development of agroecological practices. To this end, the SAS team is developing two lines of research: the first is based on a non-targeted approach of association genetics and metabolomics of root exudates, the second aims at characterizing a family of receptors for allelopathic compounds, the D14/KAI2 proteins.
Genes of interest potentially involved in intra-specific allelopathy
Through the ALLELO project (2019-2021) funded by the Carnot Institute Plant2Pro, the SAS team, in collaboration with IGEPP Rennes and LIPME in Toulouse, has set up an innovative system for phenotyping allopathic properties under controlled conditions, called "Plant Soil Feedback (PSF)", which makes it possible to avoid the effects of competition. Combining analyses with this system with association genetics in the model plant Arabidopsis thaliana, and the use of a Phenoscope in the IJPB Plant Observatory, about fifteen candidate genes potentially involved in intra-specific allelopathy have been identified. Two of these are involved in the metabolism of glucosinolates, a large family of sulfur heterosides, specific to the Brassicales order. These metabolites are actors of plant defense and are known to be exuded/released by roots in Arabidopsis and may have allelopathic properties. Their role remains to be validated before transfer to agronomic applications with cultivated species, such as rapeseed (a species of major agronomic interest) or camelina. The validation of these candidate genes is the subject of a thesis financed 100% by the INRAE BAP division starting in October 2022. Its objective is to validate the involvement of glucosinolates in allelopathy, as well as to identify the exact type of glucosinolates or derivatives responsible for these effects. In addition, the PlantAlliance consortium is also supporting work in the team by funding the ALLOGA project which is at the interface between genetics and agronomy. This project, conducted in collaboration with the IGEPP and Agroecology units in Rennes and Dijon, respectively, aims to adapt the PSF experimental system developed on Arabidopsis to rapeseed in order to evaluate the allelopathic properties of a wide range of rapeseed varieties. The knowledge obtained will make it possible to integrate allelopathy into a mechanistic model and to thereby quantify the effects of cropping systems on the dynamics of weed flora. These data could be used to evaluate the allelopathic properties of rapeseed in cultivation.
"Multi-faceted" allelopathic signal receptors
The second aspect of SAS team's research is based on knowledge acquired about the D14/KAI2 proteins which are strigolactone receptors. The work of the team on different species such as Pisum sativum (pea), Arabidopsis thaliana, Physcomitrium patens (moss) or the parasitic plants Phelipanche ramosa, and Orobanche cumana allowed us to demonstrate that these proteins are involved in the perception of other allelopathic compounds. Recently, we demonstrated that PpKAI2 proteins (11 in number) from the non-vascular plant, P. patens, were involved in the development of this moss and the perception of allelopathic signals (Lopez Obando 20211, Guillory & Bonhomme 20222). In order to identify the allelopathic signal(s) perceived by these proteins, the SAS team in collaboration with the” Institut de Chimie des Substances Naturelles” has now developed probes and molecules that allow a better understanding of their function and has established ligand identification strategies (de Saint Germain et al 20223, Fornier et al 20224). Determination of the crystallographic structure of two pea KAI2 proteins, in collaboration with UC Davies University, revealed a duplication event in KAI2 proteins in legumes that confers the ability to perceive a greater diversity of rhizosphere signals (Guercio et al 20225). 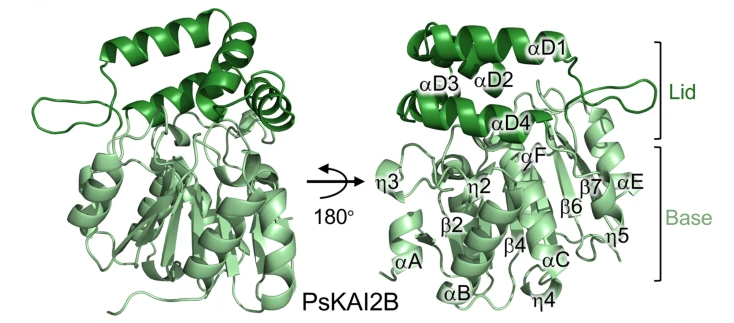
Overview of PsKAI2B structure. Lid and base domains are colored in forest and light green respectively with secondary structure elements labeled.
The SAS team is working at the international level to push back scientific forefronts, the work of the SAS team using an integrated approach that combines genetics, phytochemistry, physiology and agronomy to better understand the genetic and molecular determinants of allelopathy towards developing allelopathic varieties in crops for biological weed control.
In September 2022, Sophie Jasinski (research Scientist) and Alain Lécureuil (Technician) will join the team.
Back
Scientific Highlight of the INRAE BAP division
"Strigolactones and Allelochemicals Signaling" team SAS
Associated publications
> de Saint Germain A, Clavé G, Schouveiler p, Pillot JP , Singh A-V, Chevalier A, Suzanne Daignan Fornier, Ambre Guillory, Sandrine Bonhomme, Catherine Rameau, Boyer F-D, Expansion of the Strigolactone Profluorescent Probes Repertory: The Right Probe for the Right Application, Front Plant Sci https://doi.org/10.3389/fpls.2022.887347 (2022)
> Daignan Fornier S, de Saint Germain A, Retailleau P, Pillot J-P, Quentin Taulera Q,Andna L, L Miesch L, Rochange S, Pouvreau J-B, Boyer F-D, Noncanonical Strigolactone Analogues Highlight Selectivity for Stimulating Germination in Two Phelipanche ramosa Populations, Nat Prod https://doi.org/10.1021/acs.jnatprod.2c00282 (2022)