Semi-automated quantification of phosphorylation signals of a histone involved in cell division in Arabidopsis
Cell biology
3D Image analysis
3D signal quantification
Novel 3D microscopy image analysis tool: research from the SPACE and MIN teams published in New Phytologist
Cell division is regulated by numerous post-translational modifications of histones. Among these, phosphorylation of histone H3 at serine 10 is particularly critical for ensuring accurate chromosome segregation during mitosis. This study aimed to precisely characterise the dynamics of this phosphorylation during mitosis in plant cells, in order to better understand the underlying molecular pathways.
To achieve this, a 3D image analysis pipeline was developed based on the immunolocalisation of H3S10ph, enabling semi-automated quantification of H3S10 phosphorylation in mitotic cells of Arabidopsis roots. Additionally, a novel method was established to correct for signal attenuation with image depth, a common limitation of 3D imaging. This approach relies on directly measuring the intensity of objects of interest and quantifying their attenuation through the image stack. By accounting for the signal attenuation profile, this correction significantly enhances both the precision and statistical robustness of the analyses. Using this pipeline, subtle differences in H3S10ph levels were observed in mutants with altered PP2A phosphatase activity, suggesting an indirect role of PP2A enzymes in regulating H3S10ph levels.
This tool provides new opportunities to study these regulatory pathways in plants, taking advantage of the extensive genetic resources available in Arabidopsis. It can also be adapted to investigate other post-translational histone modifications or, more generally, any discrete 3D signal.”
Research developed at the Institute Jean-Pierre Bourgin for Plant Sciences in collaboration.
To achieve this, a 3D image analysis pipeline was developed based on the immunolocalisation of H3S10ph, enabling semi-automated quantification of H3S10 phosphorylation in mitotic cells of Arabidopsis roots. Additionally, a novel method was established to correct for signal attenuation with image depth, a common limitation of 3D imaging. This approach relies on directly measuring the intensity of objects of interest and quantifying their attenuation through the image stack. By accounting for the signal attenuation profile, this correction significantly enhances both the precision and statistical robustness of the analyses. Using this pipeline, subtle differences in H3S10ph levels were observed in mutants with altered PP2A phosphatase activity, suggesting an indirect role of PP2A enzymes in regulating H3S10ph levels.
This tool provides new opportunities to study these regulatory pathways in plants, taking advantage of the extensive genetic resources available in Arabidopsis. It can also be adapted to investigate other post-translational histone modifications or, more generally, any discrete 3D signal.”
Research developed at the Institute Jean-Pierre Bourgin for Plant Sciences in collaboration.
Back
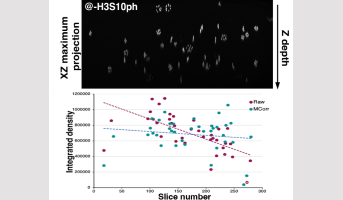
Legend: Top, an XZ projection of a 3D root image stack, following immunolocalisation, showing the decrease in H3S10 signal intensity with depth (Z). Bottom, a graph illustrating the correction of this signal attenuation using the pipeline developed in this study (difference in slope between MCorr and raw data)
IJPB highlight
Reference
Kelemen A, Uyttewaal M, Máthé C, Andrey P, Bouchez D, Pastuglia M. Semiautomatic quantification of 3D Histone H3 phosphorylation signals during cell division in Arabidopsis root meristems. New Phytol 2025. doi: 10.1111/nph.70365.
Contact : Martine Pastuglia
IJPB teams
> Spatial Control of Cell Division SPACE
> Modeling and Digital Imaging MiN
Collaborating team
Department of Botany, Faculty of Science and Technology, University of Debrecen, Debrecen, Hungary
IJPB highlight
Reference
Kelemen A, Uyttewaal M, Máthé C, Andrey P, Bouchez D, Pastuglia M. Semiautomatic quantification of 3D Histone H3 phosphorylation signals during cell division in Arabidopsis root meristems. New Phytol 2025. doi: 10.1111/nph.70365.
Contact : Martine Pastuglia
IJPB teams
> Spatial Control of Cell Division SPACE
> Modeling and Digital Imaging MiN
Collaborating team
Department of Botany, Faculty of Science and Technology, University of Debrecen, Debrecen, Hungary