Identification of genes and specialized metabolites involved in allelopathy (Alexandre de Saint Germain, Sophie Jasinski)
Mechanism of strigolactone perception (Alexandre de Saint Germain)
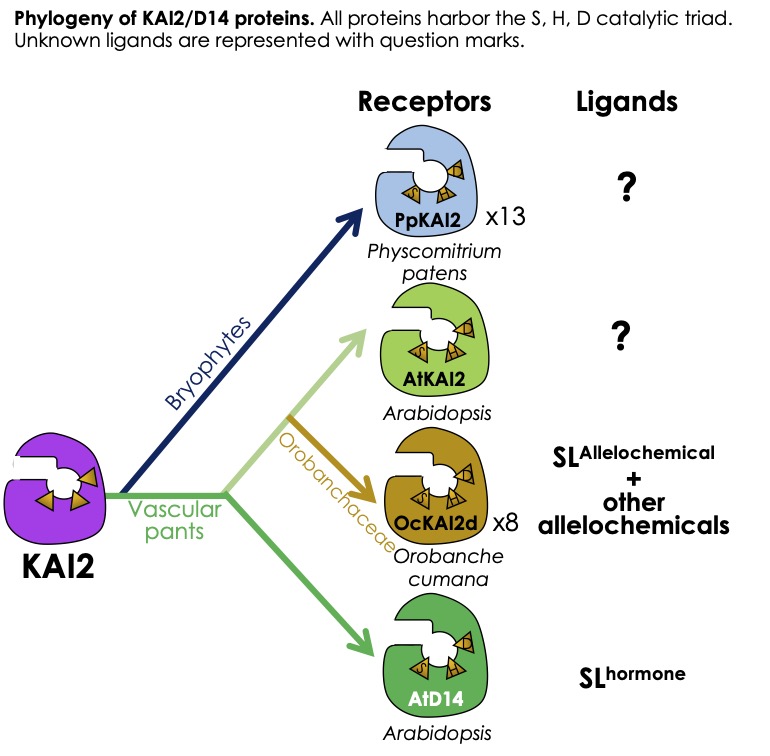
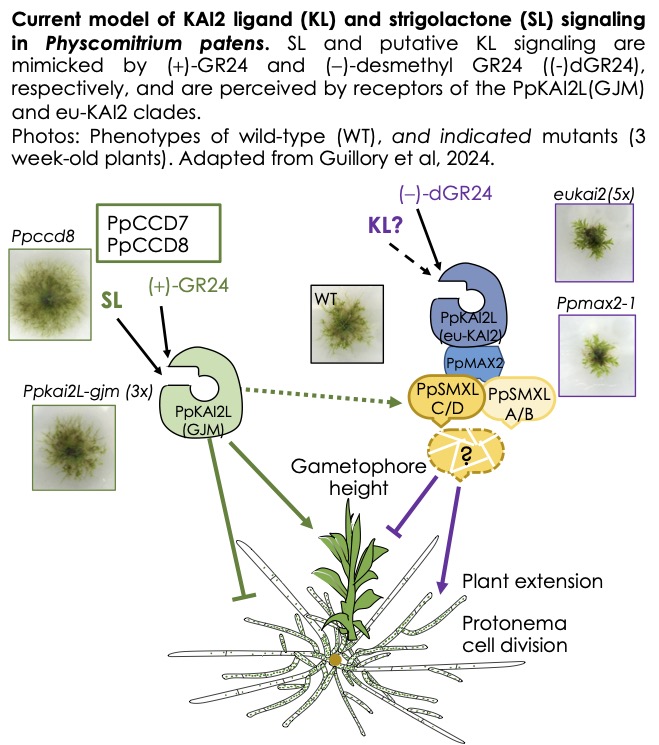
Evolution of strigolactone signalling (Sandrine Bonhomme)
Control of shoot branching (Catherine Rameau, Alexandre de Saint Germain)
Our agriculture is facing the challenge of agro-ecological transition, with the need to progressively reduce the use of herbicides. The development of innovative strategies for tomorrow's agriculture could come from new knowledge on the natural mechanisms of regulation of plant-plant interactions, such as allelopathy. This process by which plants release chemical compounds into the rhizosphere, molecules that modify the growth of neighbouring plants, appears to be an interesting lever for promoting the biological regulation of weeds. To date, few allelopathic compounds have been identified and our knowledge of the molecular mechanisms involved is limited, partly due to methodological constraints. The SAS team is developing projects combining genetic and metabolomic approaches to identify the genes and metabolites involved in allelopathy. Using an innovative phenotyping device for allelopathic properties and association genomics studies (GWAS), it has been possible to identify candidate genes potentially involved in the biosynthesis of allelopathic signals in the model plant Arabidopsis.


Mechanism of strigolactone perception (Alexandre de Saint Germain)
Strigolactones (SL) are perceived as plant hormones by land plants, as germination stimulants by parasitic plants of the family Orobanchaceae (genera Striga, Orobanche, Phelipanche), and as host presence signals by arbuscular endomycorrhizal fungi. The strigolactone receptor in vascular plants (D14 in rice, RMS3 in pea) is a member of the large family of α/β-hydrolases. These proteins are both receptors and enzymes. After characterizing the original mechanism by which the SL receptor carries out an irreversible enzymatic reaction to generate its own ligand, the team turned to the KAI2 proteins, which are homologous to D14. Characterization of kai2 mutants from several species suggests that these proteins perceive allelochemicals from the rhizosphere, such as karrikins, but also one (or more) as yet unknown endogenous compound(s). In obligate parasitic plants such as Orobanche cumana, a sunflower parasite, we observed (as in the moss P. patens, see below) a significant diversification of KAI2 proteins, suggesting a diversity of perceived compounds. The team is developing projects combining genetic, biochemical and chemical approaches to identify the ligands of these KAI2 proteins and describe the associated perception mechanisms.

Evolution of strigolactone signalling (Sandrine Bonhomme)
A recent study on the bryophyte Marchantia paleacea suggests that in the first land plants, SLs did not have a hormonal role but rather a role in the rhizosphere, in particular for the establishment of symbiosis with mycorrhizal fungi. In the model bryophyte used in the laboratory, the moss Physcomitrium patens, SLs appear to have a hormonal role (regulating development) and an allelopathic role, as they are exuded by plants and regulate the spread of neighbouring plants. The team has identified numerous homologs of SL receptors in P. patens (PpKAI2L proteins) and is seeking to gain a better understanding of the evolution of signalling pathways for these molecules, using molecular genetic approaches. CRISPR-Cas9 technology, which enables multiple gene editing in P. patens, is being used to generate mutants and lines that over-express proteins. At the same time, the team is seeking to identify the structure of allelopathic molecules in P. patens (SLs and KAI2 ligands).

Control of shoot branching (Catherine Rameau, Alexandre de Saint Germain)
In 2008, SLs became a new class of plant hormones for their role in controlling branching, a discovery in which the team played a part. Despite Catherine Rameau's retirement in April 2024, the team will continue to work on the control of pea branching, in particular through collaborations already in place with Christine Beveridge (Queensland University, Australia). One of the current issues is the identification of the SLs that inhibit branching in pea.Each plant species produces a mixture of different SLs, and not all the genes involved in the diversification of these molecules (downstream of the biosynthesis pathway) are known. Very recently, the team embarked on a project to edit the pea genome, in collaboration with the DRAGON team (Fabien Nogué).

Leader:
Sandrine Bonhomme