Development of the RBA model to predict the plant cell response in complex environmental conditions
As part of the INRAE DIGIT-BIO metaprogram (Digital Biology for Exploring and Predicting Life), the PlantRBA exploratory project (2021 - 2023) aimed to develop, calibrate and experimentally validate a mathematical model predicting the metabolic behavior of Arabidopsis thaliana under abiotic constraints (limited water and/or nitrogen availability). This model is based on the parsimonious distribution of resources between the plant's various biological functions, thus reconciling the finest scales (genes, metabolites) with the phenotype.
Predicting the plant cell response in complex environmental conditions is a challenge in plant biology. The MaIAGE, PHYGERM, SATURNE and VAST INRAE teams developed a resource allocation model at cellular and molecular scales for the leaf photosynthetic cell of A. thaliana, based on the Resource Balance Analysis (RBA) constraint-based modeling framework. The RBA model contains the metabolic network and the major macromolecular processes involved in the plant cell growth and survival and localized in cellular compartments. The scientists simulated the model for varying environmental conditions of temperature, irradiance, partial pressure of CO2 and O2, and compared RBA predictions to known resource distributions and quantitative phenotypic traits such as the relative growth rate, the C/N ratio, and finally to the empirical characteristics of CO2 fixation given by the well-established Farquhar model. In comparison to other standard constraint-based modeling methods like Flux Balance Analysis, the RBA model makes accurate quantitative predictions without the need for empirical constraints. Altogether, the results show that RBA significantly improves the autonomous prediction of plant cell phenotypes in complex environmental conditions, and provides mechanistic links between the genotype and the phenotype of the plant cell. They have been published in the Metabolic Engineering scientific journal.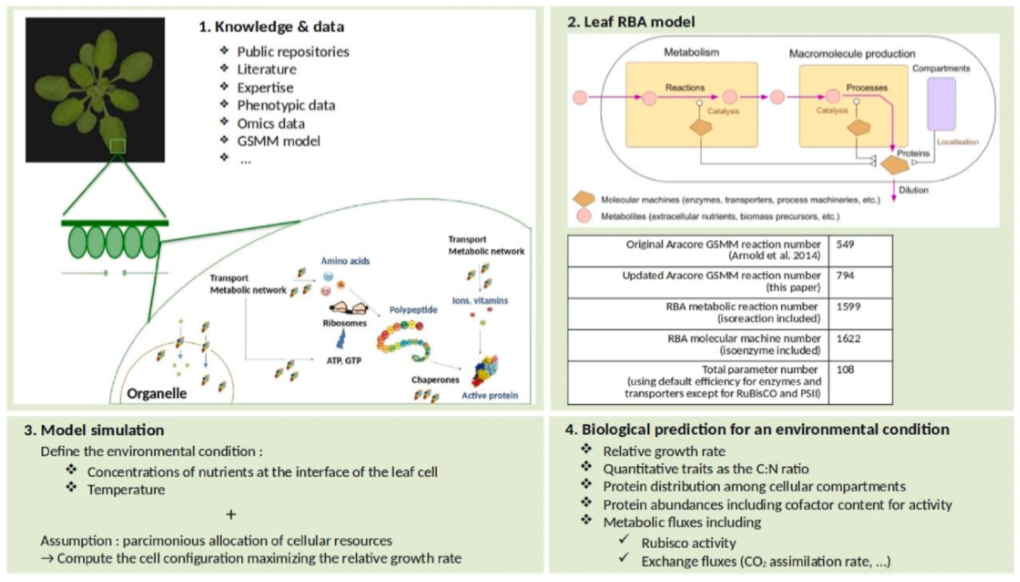
Principle of RBA model generation and simulation (Figure from Goelzer et al., Metabolic Engineering, 2024)
Research is currently being pursued through the ANR PRC collaborative research project ModLsys (2023-2028), to establish -and share with the community- a multi-scale modeling framework for living systems.
Research developed at the Institute Jean-Pierre Bourgin for Plant Sciences
Back
Principle of RBA model generation and simulation (Figure de Goelzer et al., Metabolic Engineering, 2024)
Zoom
Référence :
A Goelzer, L Rajjou, F Chardon, O Loudet, V Fromion. (2024). Resource allocation modeling for autonomous prediction of plant cell phenotypes. Metabolic Engineering, doi : https://doi.org/10.1016/j.ymben.2024.03.009
Contact :
Olivier Loudet, contact
IJPB teams :
> "Germination Physiology" PHYGERM
> "Senescence, Autophagy, Nutrient Recycling and Nitrogen Use Efficiency" SATURNE
> "Variation and Abiotic Stress Tolerance " VAST
Collaborating team :
"Applied Mathematics and Informatics from Genome to the Environment" MaIAGE