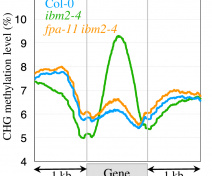
In plants, DNA methylation, that occurs at cytosines in all contexts, is an epigenetic mark critical for transposon silencing. Our group studies the different molecular pathways controlling DNA methylation and its inheritance over generations.
We use two model plants differently enriched in transposable elements: tomato and Arabidopsis.
Biological Question
In plants, DNA methylation occurs in all cytosine contexts, mainly to silence repeats and transposons found in heterochromatic regions.
Epialleles, identified in different organisms and predominantly in plants, are gene variants based on epigenetic marks stably transmitted between generations. Most of the plant epialleles described so far depend on DNA methylation of cytosines, an epigenetic mark influencing the way genes are transcribed and a hallmark of transposon silencing. We aim at understanding the extent of epigenetic variation, whether induced or natural to better described the regulations governing DNA methylation homeostasis in plants.
Models, tools and methods
Our model plants are both Arabidopsis and tomato. We are working with plants carrying modified epigenomes and we address questions of basic science using whole-genome bisulfite sequencing technologies and bioinformatics, associated with conventional molecular biology tools.
Societal and economical impacts
Our research will help to enhance plant breeding by deciphering new ways to control the epigenetic diversity within cultivated populations.
Biological Question
In plants, DNA methylation occurs in all cytosine contexts, mainly to silence repeats and transposons found in heterochromatic regions.
Epialleles, identified in different organisms and predominantly in plants, are gene variants based on epigenetic marks stably transmitted between generations. Most of the plant epialleles described so far depend on DNA methylation of cytosines, an epigenetic mark influencing the way genes are transcribed and a hallmark of transposon silencing. We aim at understanding the extent of epigenetic variation, whether induced or natural to better described the regulations governing DNA methylation homeostasis in plants.
Models, tools and methods
Our model plants are both Arabidopsis and tomato. We are working with plants carrying modified epigenomes and we address questions of basic science using whole-genome bisulfite sequencing technologies and bioinformatics, associated with conventional molecular biology tools.
Societal and economical impacts
Our research will help to enhance plant breeding by deciphering new ways to control the epigenetic diversity within cultivated populations.

Leader:
Nicolas Bouché