The team SYNAPS dedicates its research on finding solutions that favor the agroecological transition. More specifically, the team’s major objective is to understand the molecular and physiological mechanisms improving crop Nitrogen Use Efficiency (NUE) mediated by beneficial microorganisms.
Welcome to the SYNAPS team webpage!

Scientific goals :
The objective of the research program is to improve or maintain plant productivity when nitrogen fertilization is reduced. To achieve this, we exploit the beneficial impact of the microbial environment in the rhizosphere and the existing or created genetic diversity of crops with regard to nitrogen uptake and assimilation (Figure 1).
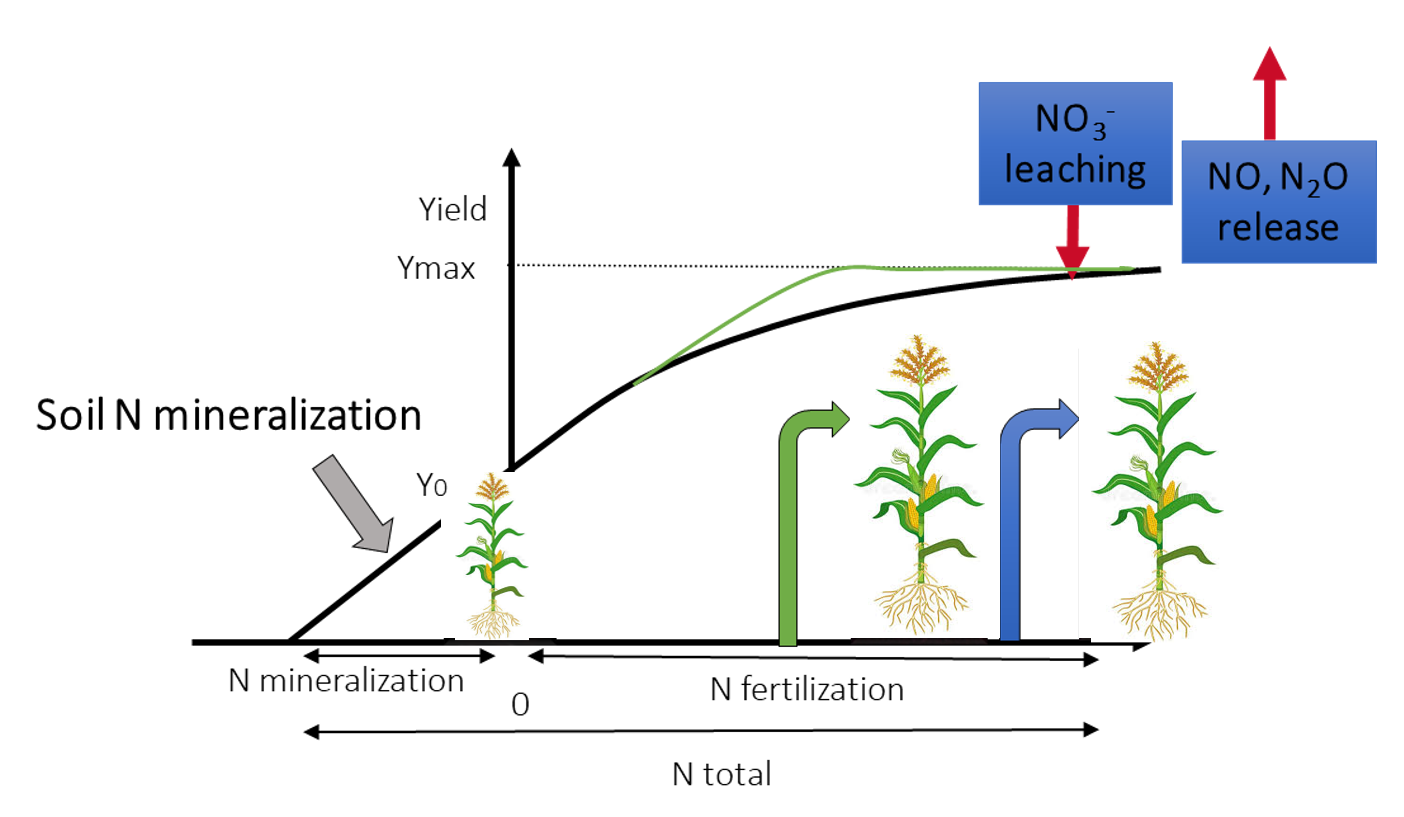
Figure 1: Nitrogen use efficiency (NUE) allows determining the capacity of a crop plant to manage the available N to produce gran yield or biomass yield. The plant with the green arrow has a better NUE than the one with the blue arrow. Higher NUE leads to less fertilizer use and less environmental impact.
Several levers can improve crop NUE including breeding, rotations, intercropping and exploiting microbiome to increase N availability. The team implements a systems biology approach, using crop models, integrating multi-omics, genomics, metabolic modelling and microbial ecology to characterise plant N nutrition mediated by its microbiome. Our research is focused on cereals such as maize and sorghum alone or intercropping with legumes such as soybean (Figure 2) .
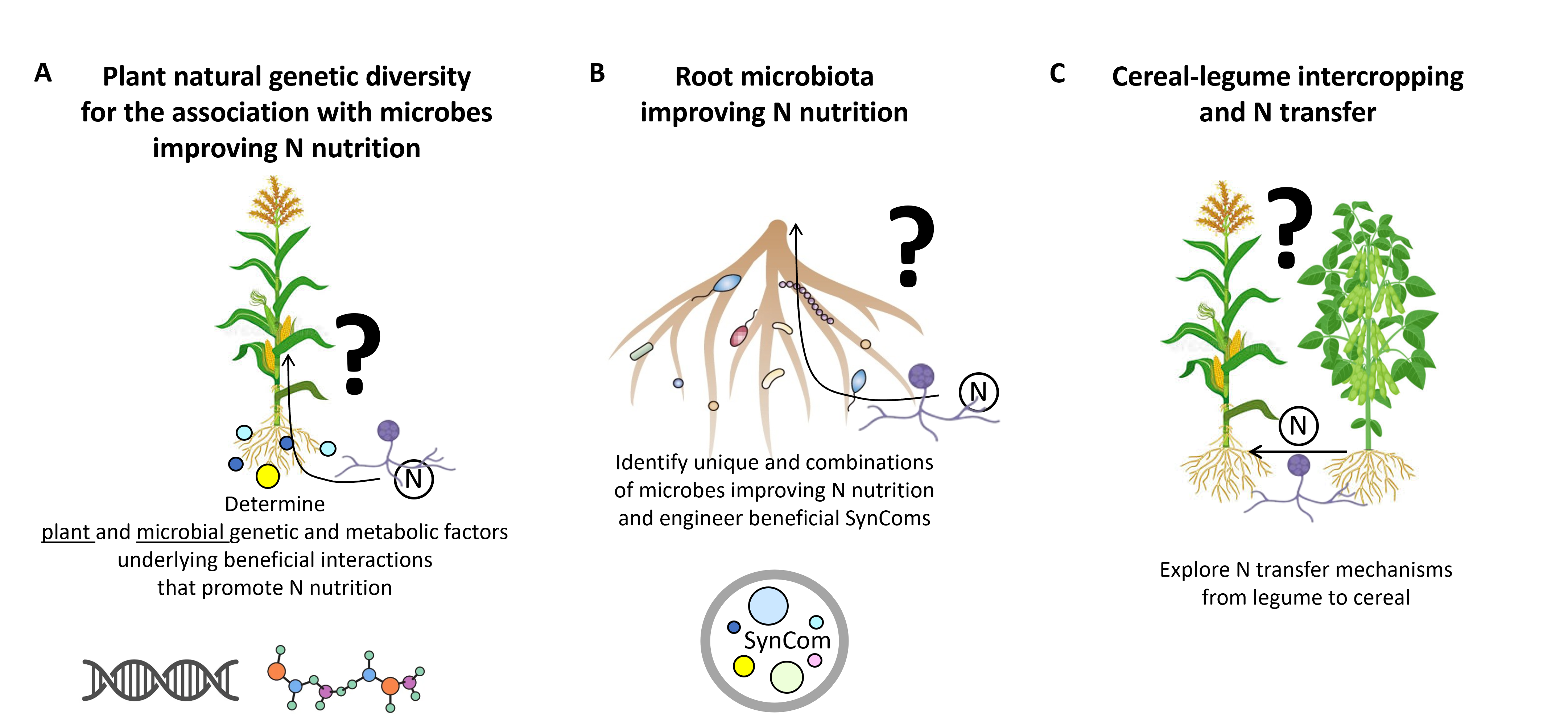
Figure 2: Three main axes of research for sustainably improved N use. A) Understanding the genetic, physiological and metabolic mechanisms taking place in the plant and in the microbes that improve N use. B) Engineering crop microbiome by creating Synthetic Communities (SynComs) based on their metabolic complementarity in boosting AMF mediated N crop nutrition. C) Understanding the genetic, physiological and metabolic mechanisms taking place in the intercropping systems and the role of the microbiome in favoring the N transfer via mycorrhizal network connecting cereals to legumes.
Our biological questions:-
A) What are the physiological, metabolic and molecular mechanisms involved in maize N nutrition during symbiosis with AMF? We demonstrated that maize interaction with the arbuscular mycorrhizal fungus (AMF) Rhizophagus irregularis alleviates maize N deficiency stress (Figure 3) and are developing a series of biological and experimental tools to understand by which mechanisms this process takes place (Collaboration with D. WIPF and P-E. COURTY). We develop and integrate multi-omics approaches, including transcriptomic, metabolomic, fluxomic, and ionomics data at the level of plant organs and including the microbial compartments. Using microscopy we track the fate of arbuscules during symbiosis (Figure 4) trying to determine whether their decay may provide N to the plant. These approaches will help us to better understand the physiological and molecular processes underlying maize-microbiota interactions which improve maize nitrogen use efficiency.
 Figure 3: Maize plants grown under limiting N and inoculated with the AMF R. irregularis (right) or not (left).
Figure 3: Maize plants grown under limiting N and inoculated with the AMF R. irregularis (right) or not (left).We implemented system biology approaches based on multi-omics analysis and metabolic modeling tools. Owing to a collaboration with Dr Rajib SAHA (Univ. Nebraska) our study supports the idea that the pyrimidine metabolic pathway is a crucial component of maize nitrogen metabolism during AM symbiosis under nitrogen-limited conditions (Decouard et al., 2023, BioRxiv, Chowdhury et al. 2023). In collaboration with the IJPB team NUTS, we are investigating the response of Brachypodium distachyon to R. irregularis under low N condition. In collaboration with the IJPB team QALIBIOSEC we are investigating the role of plant cell walls in AM symbiosis and vice versa.
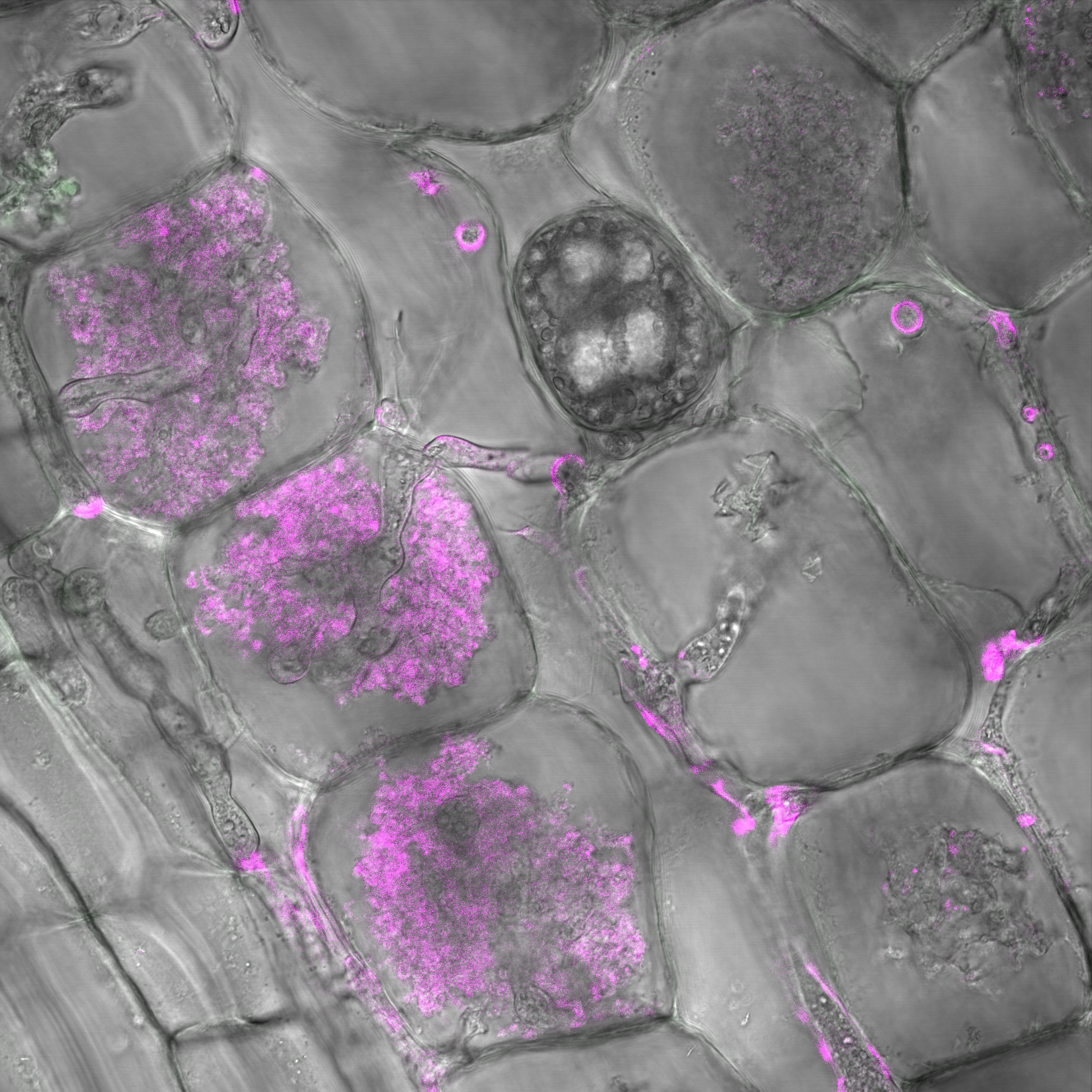 Figure 4: Maize roots colonized by arbuscules stained with wheat germ agglutinin (WGA). Pink structures are AMF arbuscules and hyphae.
Figure 4: Maize roots colonized by arbuscules stained with wheat germ agglutinin (WGA). Pink structures are AMF arbuscules and hyphae.B) What are the microbial consortia that can boost maize N nutrition via AMF and by which mechanism ? This research will delve into the mechanisms of maize microbiota recruitment by root exudates and the characterization of the microbial consortia associated with AMF-associated maize root boosting nitrogen nutrition. To address these points, we developed an original aeroponics based maize-AMF cocultivation system (Figure 5) to study what metabolites secreted in the plant root exudates may play a role during mycorrhizal symbiosis (Decouard et al. 2025 JXB. in press).
 Figure 5: Maize-AMF co-cultivation aeroponics based system. This system allows studying root exudate metabolic composition.
Figure 5: Maize-AMF co-cultivation aeroponics based system. This system allows studying root exudate metabolic composition. We will especially focus our research on rhizospheric free-living and endophytic nitrogen-fixing bacteria associated with maize roots and mycorrhizal hyphosphere. This work will be implemented in three major steps: trapping bacteria and fungi in the rhizosphere (collab J. Fievet, GQE), creating synthetic communities (Syncom), determining the exudate compounds involved in their recruitment and optimizing the synergistic effect of the SynCom-AMF combination on N maize metabolism. The impact of the bacterial consortia and AMF on maize N metabolism will be characterized.

Figure 6: Systems to trap bacteria associated with the rhizosphere and hyphosphere in a maize-AMF cocultivation soil based system.
C) What are the mechanisms involved in N nutrition under legumes and cereals intercropping system in presence of both AMF and N-fixing legume symbionts ? Intercropping systems represent key agroecological strategies to optimize and reduce the use of synthetic nitrogen fertilizers (Figure 7). We aim to explore the beneficial effect of the Common Mycorrhizal Network in redistributing the legumes/rhizobium-derived biologically fixed nitrogen towards cereals as well as to explore the impact of the presence of the legume/rhizobium couple on maize primary and secondary metabolisms under such a continuum of interaction involving AMF.
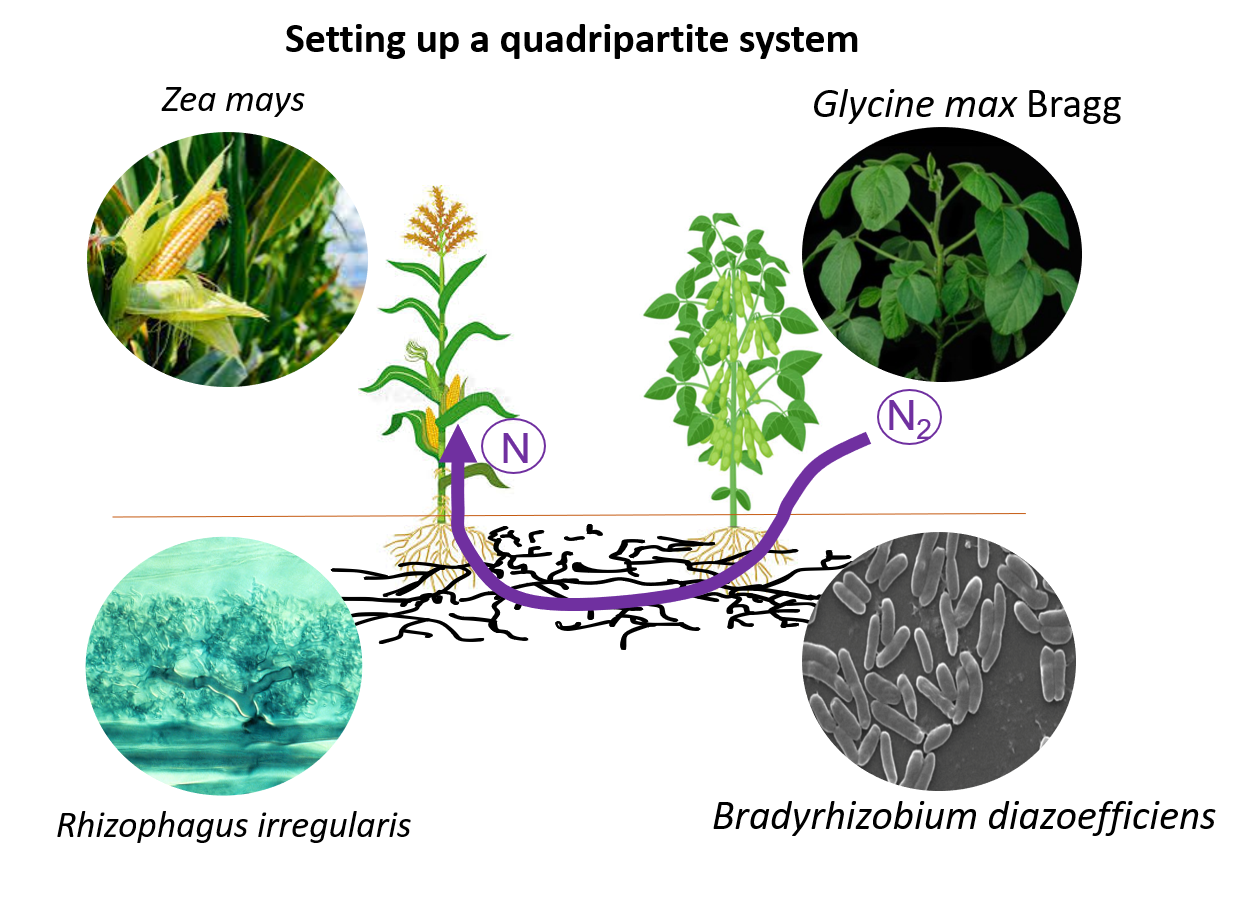
Figure 7: Maize-AMF-soybean-Bradyrhizobium quadripartite system for mechanisms of intercropping mediated N crop metabolic mechanisms.
We use the cereal crop maize or sorghum, the legume crop soybean, the arbuscular mycorrhizal fungus Rhizophagus irregularis and the nitrogen-fixing bacterium strain Bradyrhizobium diazoefficiens in the greenhouse (Figure 8) and in the field. The role of the microbiome on such an intercropping system will be investigated.
Figure 8: Maize-soybean intercropping systems in the greenhouse.
Models, tools and methods
The main biological models are maize, sorghum, soybean for the plant, and for the microorganisms, we use the AMF R. irregularis, and the nitrogen fixing bacterium Bradyrhizobium diazoefficiens. Isolated microorganisms are still to be characterized in a collection. Several methodological approaches are used on different organs and at different scales of plant organization and on the microbial partners.
These are our approches:
-Physiology, metabolism and molecular genetics of nitrogen nutrition.
-Microbiology, microbial ecology, culturomics.-Metabarcoding, metagenomics.
-Phenotyping from cellular to whole plant level under controlled or field conditions.
-Systems biology, multi-omics (transcriptomics, metabolomics, proteomics).
-Metabolic modeling.
Economic and societal issues
The research conducted by the SYNAPS team consists of providing tools and markers to select cultivated plants that use nitrogen more efficiently, thus reducing the excessive use of nitrogen fertilizers that are harmful to the environment. As such, the SYNAPS team is involved in innovative projects involving industrial partners and technical institutes.
Major recent publications (to see morefollow this link):
Smith NT, Boukherissa A, Antaya K, Howe GW, Mergaert P, Rodríguez de la Vega RC, Shykoff JA, Alunni B, diCenzo GC (2025). Taxonomic distribution of SbmA/BacA and BacA-like antimicrobial peptide transporters suggests independent recruitment and convergent evolution in host-microbe interactions. Microb Genom, 11(4):001380. PubMed | DOI
Urrutia M, Blein-Nicolas M, Fernandez O, Bernillon S, Maucourt M, Deborde C, Balliau T, Rabier D, Benard C, Prigent S, Quilleré I, Jacob D, Gibon Y, Zivy M, Giauffret C, Hirel B, Moing A (2024). Identification of metabolic and protein markers representative of the impact of mild nitrogen deficit on agronomic performance of maize hybrids. Metabolomics, 20(6):128. PubMed | DOI
Chowdhury NB, Simons-Senftle M, Decouard B, Quilleré I, Rigault M, Sajeevan KA, Acharya B, Chowdhury R, Hirel B, Dellagi A, Maranas CD, Saha R (2023). A multi-organ maize metabolic model connects temperature stress with energy production and reducing power generation. iScience, 26(12):108400. PubMed | DOI
Terce Laforgue T, Lothier J, Limami AM, Rouster J, Lea PJ, Hirel B (2023). The Key Role of Glutamate Dehydrogenase 2 (GDH2) in the Control of Kernel Production in Maize (Zea mays L.). Plants, 12(14):2612. PubMed | DOILimami AM, Cukier C, Hirel B (2023). 15N-labelling of Leaves Combined with GC-MS Analysis as a Tool for Monitoring the Dynamics of Nitrogen Incorporation into Amino Acids. Methods Mol Biol, 2642:151-161. PubMed | DOI
Chowdhury NB, Schroeder WL, Sarkar D, Amiour N, Quilleré I, Hirel B, Maranas CD, Saha R (2022). Dissecting the metabolic reprogramming of maize root under nitrogen-deficient stress conditions. J Exp Bot, 73(1):275-291. PubMed | DOIValderrama-Martín JM, Ortigosa F, Ávila C, Cánovas FM, Hirel B, Cantón FR, Cañas RA (2022). A revised view on the evolution of glutamine synthetase isoenzymes in plants. Plant J. PubMed | DOI
Amiour N, Décousset L, Rouster J, Quenard N, Buet C, Dubreuil P, Quilleré I, Brulé L, Cukier C, Dinant S, Sallaud C, Dubois F, Limami AM, Lea PJ, Hirel B (2021). Impacts of environmental conditions, and allelic variation of cytosolic glutamine synthetase on maize hybrid kernel production. Commun Biol, 4(1):1095. PubMed | DOIDellagi A, Quilleré I, Hirel B (2020). Beneficial soil-borne bacteria and fungi: a promising way to improve plant nitrogen acquisition. J Exp Bot, . PubMed | DOI
Dellagi A, Quilleré I, Hirel B (2020). Beneficial soil-borne bacteria and fungi: a promising way to improve plant nitrogen acquisition. J Exp Bot, . PubMed | DOI
Books:
- Philippe Reignault, Ivan Sache, Mathias Choquer, Marie-France Corio-Costet, Alia Dellagi, Frédéric Suffert Phytopathologie, (2023) ISBN-13 : 9782807302884.
- (Chapter) Guellim, A, Hirel, B., Chabrerie, O., Catterou, M., Tetu, T., Dubois, F., Ben Ahmed, H., Kichey, T. (2021) Identification of Physiological Traits Associated with Salinity and Drought Tolerance in Wheat (Triticum durum Desf.).
-(Chapter) Gorawala, P. and Mandhari, S. Eds. Agricultural Research Updates. Vol. 32, Agriculture, Imprints, Nova, Science and Technology Special Topics ISBN: 978-1-53618-974-2.
-(Chapter) Limami, A.M., Cukier, C., Hirel, B., (2023)15N-labelling of Leaves Combined with GC-MS Analysis as a Tool for Monitoring the Dynamics of Nitrogen Incorporation into Amino. In: Couée, I. (eds) Plant Abiotic Stress Signaling. Methods in Molecular Biology, vol 2642. Humana, New York, NY.
Communication visible online :
Invitation by LIED (Laboratoire Interdisciplinaire des Energies de Demain). November 2023 : https://www.youtube.com/watch?v=9gzP28d6CD4 : Title « Microbe mediated plant N nutrition »
Invitation by SNHF-SNHF (Société Nationale d’Horticulture de France). November 2020: https://www.snhf.org/revoir-le-webinaire-3-sante-des-plantes-ressources-naturelles-et-biologie-contemporaine/

Leader:
Alia DELLAGI