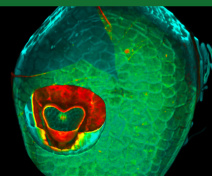
Our team studies the way plants develop and acquire their remarkable forms. To do this, we are studying the role of transcription factors that orchestrate the development using molecular genetics and imaging approaches combined with quantitative approaches and modeling.
The continuous development of plants relies on the activity of meristems. These structures contain a pool of stem cells whose maintenance and division allow generating new organs throughout the life of plants.
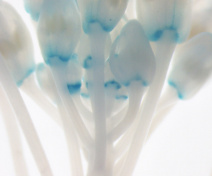
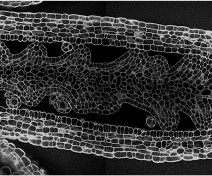
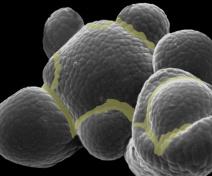
Our group is mostly interested in lateral organ boundary domains that are small group of cells separating new organs one from the other or from the meristem. These domains are also found in other organs such as in leaves or flowers. Thus, major agronomic traits including inflorescence and root architecture, organ abscission, fruit opening and seed dispersal depend on the activity of boundaries. Our work seeks to understand how boundary domains are regulated and how they impact plant development using mainly the plant model Arabidopsis thaliana.

Our group is mostly interested in lateral organ boundary domains that are small group of cells separating new organs one from the other or from the meristem. These domains are also found in other organs such as in leaves or flowers. Thus, major agronomic traits including inflorescence and root architecture, organ abscission, fruit opening and seed dispersal depend on the activity of boundaries. Our work seeks to understand how boundary domains are regulated and how they impact plant development using mainly the plant model Arabidopsis thaliana.
The aim of our group is to contribute to an integrated, multi-scale understanding of the mechanisms controlling plant morphogenesis and architecture.
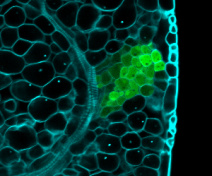
We focus on the role of boundary domains through the study of two key transcription factor families: the CUP-SHAPED COTYLEDON (CUC) family (Maugarny et al., 2015) and the three amino acid loop-extension homeodomain superfamily (TALE) (Arnaud and Pautot, 2014).
We want to further understand how these factors coordinate hormonal pathways and cellular processes such as proliferation and growth to drive plant morphogenesis, in particular leaf morphogenesis.
To link gene function with growth at the cellular and organ levels and to follow leaf morphogenesis, we developed a comprehensive set of quantitative tools.
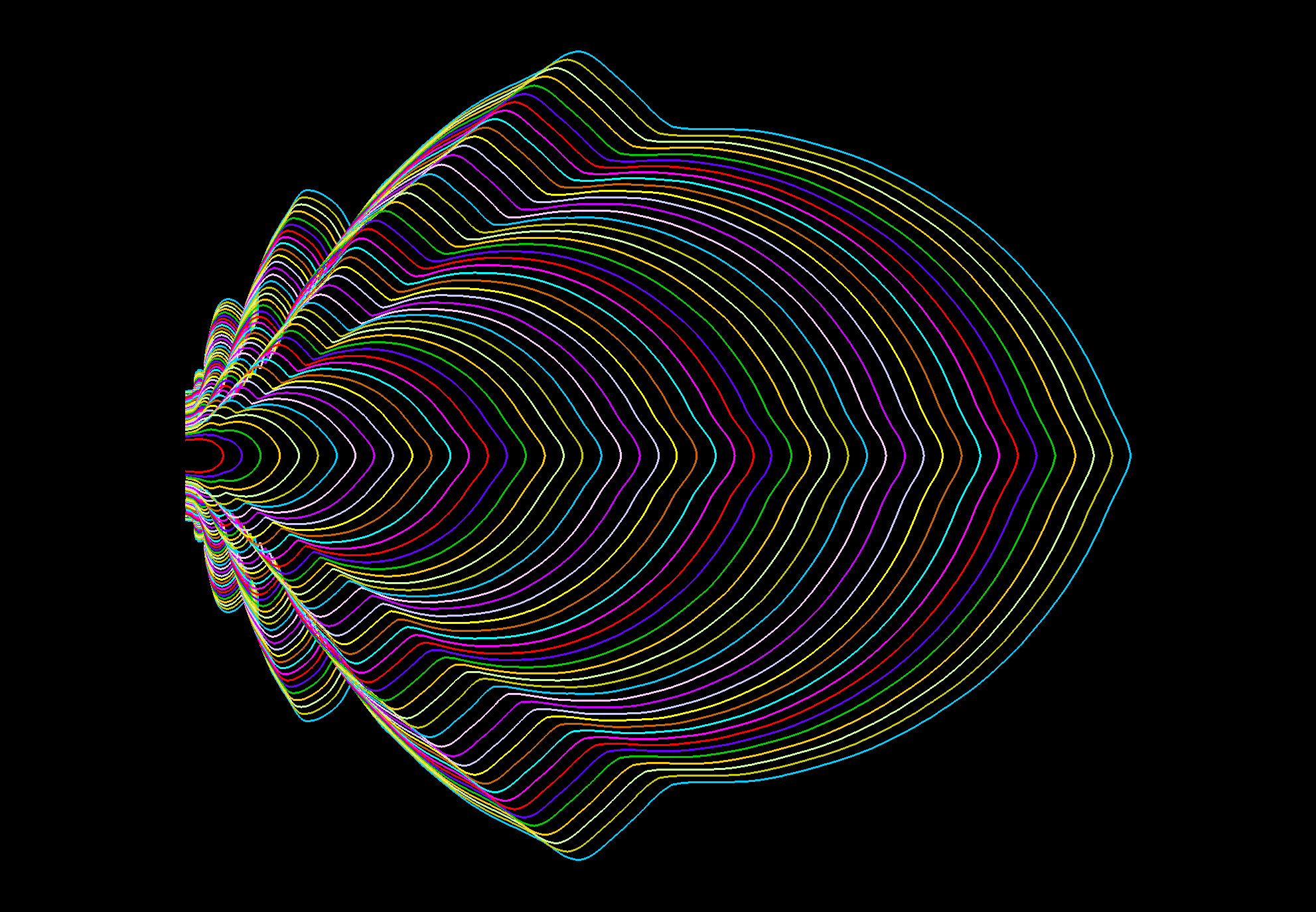
These tools are not only used to provide a multi-scale phenotyping throughout our projects, but also they fuel quantitative data to fuel the modelling approaches established through collaboration with the Modelling and Digital Imaging group at the IJPB.

These tools are not only used to provide a multi-scale phenotyping throughout our projects, but also they fuel quantitative data to fuel the modelling approaches established through collaboration with the Modelling and Digital Imaging group at the IJPB.
Besides using the leaf margin as a model, we also compare the mechanisms by which boundary domains control morphogenesis in different developmental contexts. In particular, we analyse their contribution to organ abscission and fruit dehiscence.
Through our studies we provide new understanding of the basic mechanisms driving plant development. They also allow us to put forward new innovative strategies to manipulate plant architecture in an applied perspective
Leader:
Patrick Laufs