The leaf margin, a model to understand how boundary domains control morphogenesis
- Roles of the CUC genes during leaf development
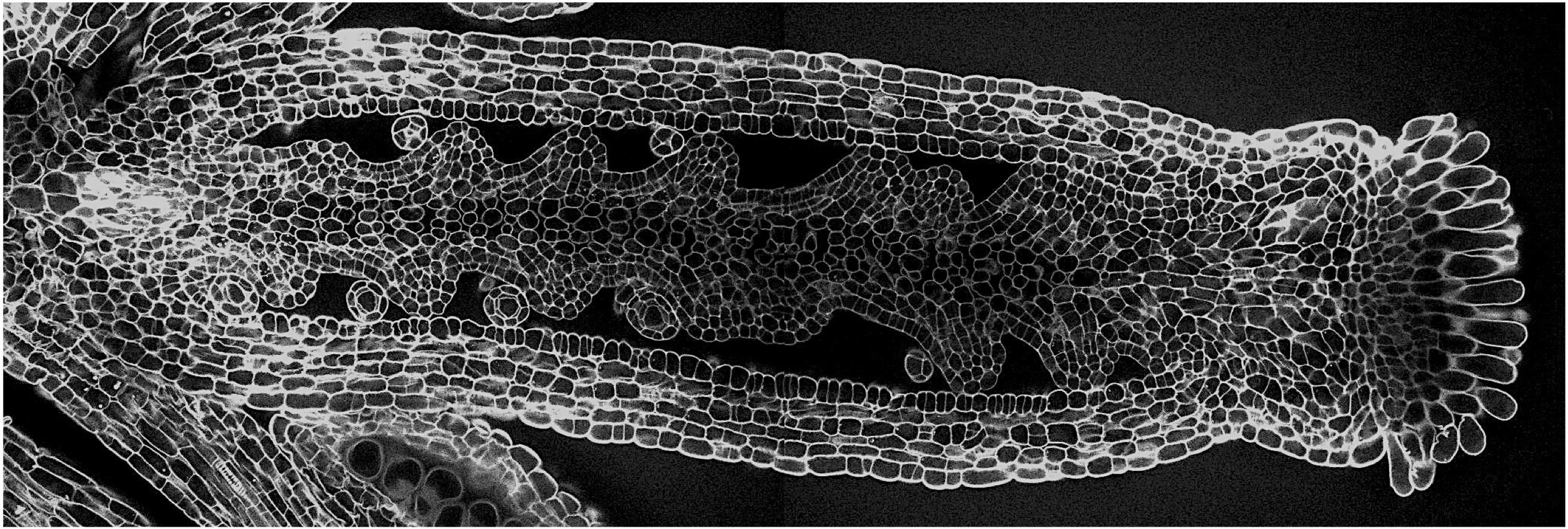
We showed previously that a balance between the CUC2 gene and its regulatory microRNA, miR164 (encoded by MIR164A) controls the level of leaf serration in Arabidopsis (Nikovics et al. 2006), thus revealing a new role for the CUC genes outside the meristem. In particular, we showed that CUC1 does not contribute to Arabidopsis leaf development, that CUC3 contributes to a later stage of leaf serration, whereas CUC2 acts during initiation of leaf serration. We also retraced the evolutionary history of CUC genes in Arabidopsis, showing that following duplication of an ancestral gene, CUC1 and CUC2 have evolved differently and acquired specific functions (Hasson et al., 2011). To extend our work in other species, we analysed the role of the CUC genes during compound leaf development in a large selection of Eudicots (Blein et al., 2008). We showed that the CUC genes have a conserved expression in the boundaries between leaflet primordia, and that they are not only required for leaflet separation but also for leaflet formation.
These observations show that the CUC genes are "universal dissectors" acting whatever the level of dissection of the leaf margin.

- Roles of the CUC genes during leaf development
We showed previously that a balance between the CUC2 gene and its regulatory microRNA, miR164 (encoded by MIR164A) controls the level of leaf serration in Arabidopsis (Nikovics et al. 2006), thus revealing a new role for the CUC genes outside the meristem. In particular, we showed that CUC1 does not contribute to Arabidopsis leaf development, that CUC3 contributes to a later stage of leaf serration, whereas CUC2 acts during initiation of leaf serration. We also retraced the evolutionary history of CUC genes in Arabidopsis, showing that following duplication of an ancestral gene, CUC1 and CUC2 have evolved differently and acquired specific functions (Hasson et al., 2011). To extend our work in other species, we analysed the role of the CUC genes during compound leaf development in a large selection of Eudicots (Blein et al., 2008). We showed that the CUC genes have a conserved expression in the boundaries between leaflet primordia, and that they are not only required for leaflet separation but also for leaflet formation.
These observations show that the CUC genes are "universal dissectors" acting whatever the level of dissection of the leaf margin.

- Exploring CUC2-dependent pathways controlling leaf shape
To identify new components of the CUC2-dependent pathway, we performed a genetic screen looking for suppressors of the strong dissection phenotype of an Arabidopsis line overexpressing CUC2.
We characterized a mutant affected in MURUS1 (MUR1), a gene coding for an enzyme responsible for GDP-L-fucose production. Our detailed morphometric analysis indicated that GDP-L-fucose is required for leaf shape, possibly via a promoting effect on CUC2 expression (Goncalves et al., 2017). Which pathway involving fucose is required to determine boundary is not yet known, but it does not appear to result from defects in xyloglucan fucosylation (Goncalves et al., 2018).
We revealed the genetic and molecular network that coordinates patterning and growth by CUC2-defined boundaries during morphogenesis (Maugarny-Calès et al., 2019). At the cellular level, CUC2 inhibits the expression of genes that control cell wall expansion, thereby stiffening the cell walls that form the boundary domains at leaf margins (Bouré et al, 2022).

To identify new components of the CUC2-dependent pathway, we performed a genetic screen looking for suppressors of the strong dissection phenotype of an Arabidopsis line overexpressing CUC2.
We characterized a mutant affected in MURUS1 (MUR1), a gene coding for an enzyme responsible for GDP-L-fucose production. Our detailed morphometric analysis indicated that GDP-L-fucose is required for leaf shape, possibly via a promoting effect on CUC2 expression (Goncalves et al., 2017). Which pathway involving fucose is required to determine boundary is not yet known, but it does not appear to result from defects in xyloglucan fucosylation (Goncalves et al., 2018).
We revealed the genetic and molecular network that coordinates patterning and growth by CUC2-defined boundaries during morphogenesis (Maugarny-Calès et al., 2019). At the cellular level, CUC2 inhibits the expression of genes that control cell wall expansion, thereby stiffening the cell walls that form the boundary domains at leaf margins (Bouré et al, 2022).

Expanding the known roles of organ boundaries during plant development.
- The known roles of boundaries during plant development have expanded during the last decade, and our group contributed to this by analysing the role of boundary domains during leaf development, inflorescence architecture, axillary meristem formation and organ abscission. We have enriched this knowledge by revealing an evolutionarily conserved role for the CUC genes during ovule separation in species with different gynoecium architectures (Goncalves et al., 2015).
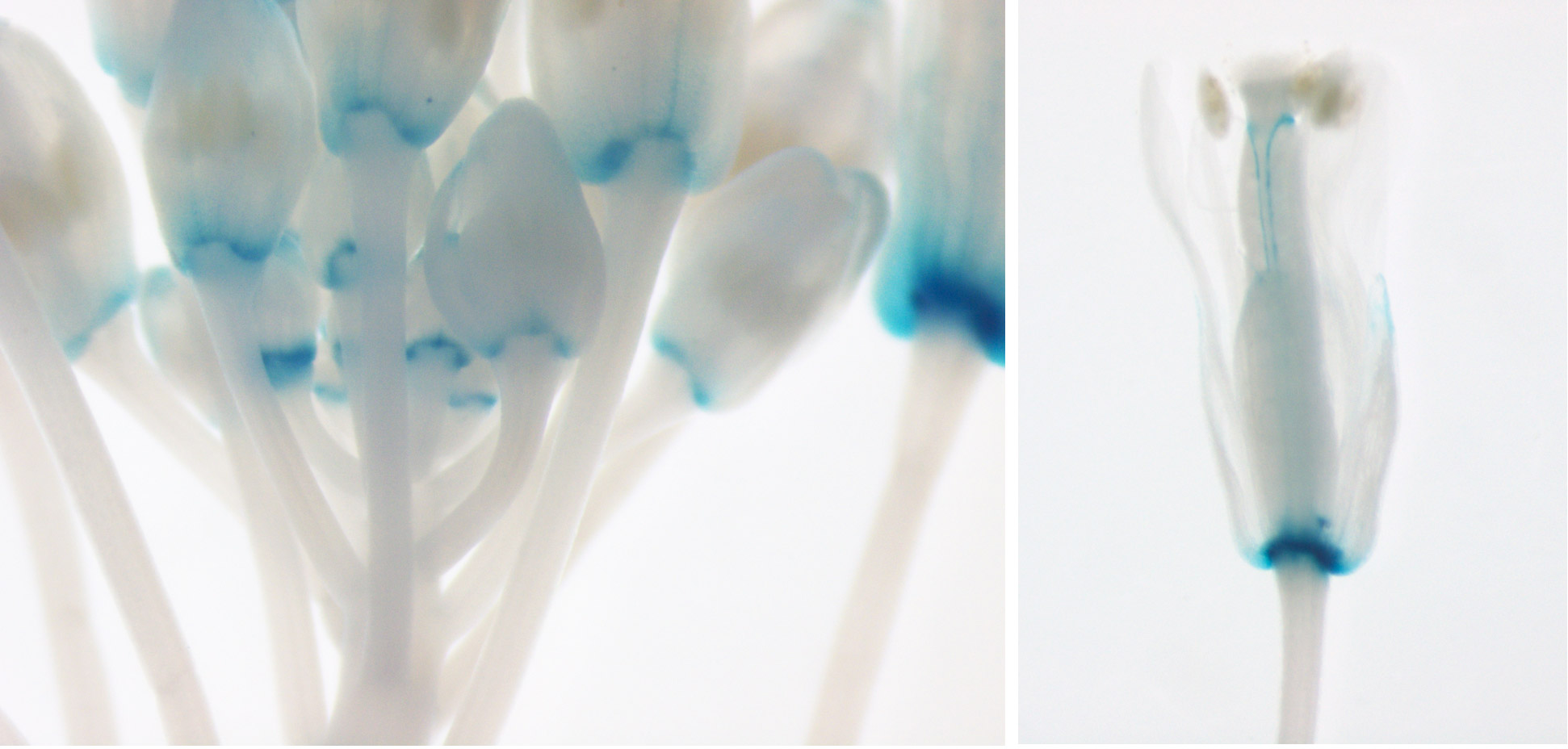
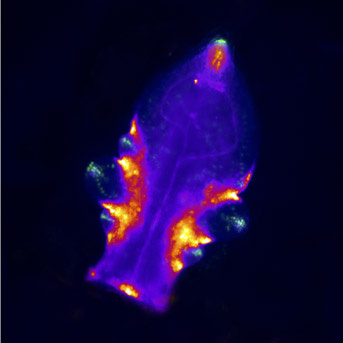
- Axillary meristem initiation also depends on CUC genes. We have shown that NGATHA-like transcription factors repress CUC expression in the meristem center to facilitate the formation of a new stem cell population (Nicolas et al., 2022).

- Axillary meristem initiation also depends on CUC genes. We have shown that NGATHA-like transcription factors repress CUC expression in the meristem center to facilitate the formation of a new stem cell population (Nicolas et al., 2022).

- The "Three amino acid loop extension” (TALE) homeodomain transcription factors constitute major regulators of meristem activity and lateral organ boundary (Hepworth and Pautot 2015). We showed that the KNAT6 member contributes to meristem and boundaries and that an antagonistic interaction between TALE members governs boundary maintenance in inflorescences (Belles-Boix et al. 2006, Ragni et al. 2008). In collaboration with the laboratory of Shelley Hepworth (Carleton University, Ottawa Canada https://carleton.ca/biology/people/shelley-hepworth/), we demonstrated that the BLADE ON PETIOLE (BOP) co-activators together with KNAT6, ATH1, KNAT2 transcription factors form a module to maintain plant boundaries (Khan et al., 2015). Currently we investigate the role of this module in two related processes depend on the activity of the boundary domains: floral organ shedding (abscission) and fruit opening (dehiscence).

Leader:
Patrick Laufs