 |
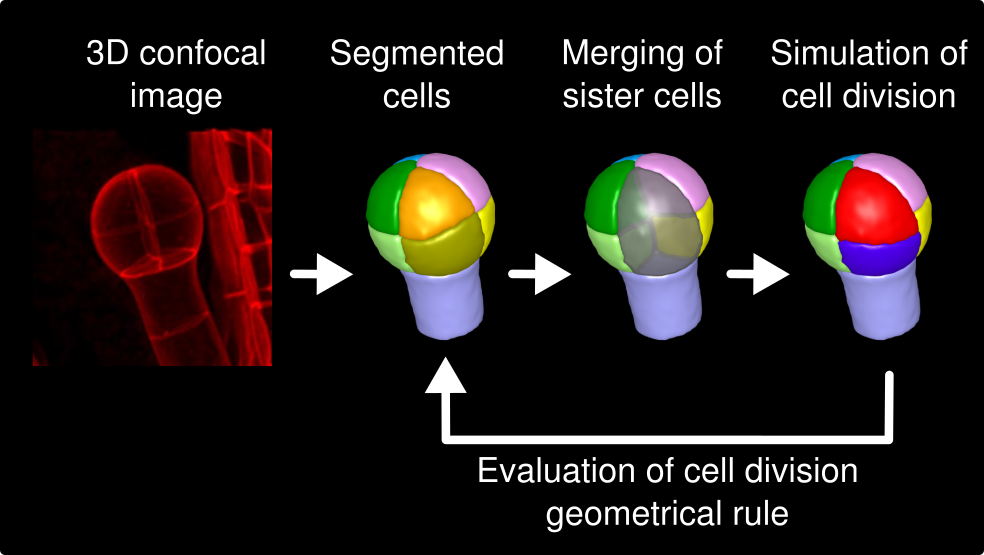
Modeling division patterns in Arabidopsis embryo (Moukhtar et al 2019, Laruelle et al 2022) : we developed a new, image-based model of cell division in 3D. Using this model, we showed that a single geometrical rule could explain the division patterns observed in Arabidopsis thaliana early embryo. |
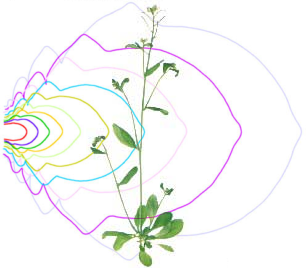 |
Quantification of morphogenetic dynamics (Biot et al 2016, Oughou et al 2023) : we developed a pipeline and a software tool to reconstruct organ growth dynamics from series of images of individual samples taken at various developmental stages. |
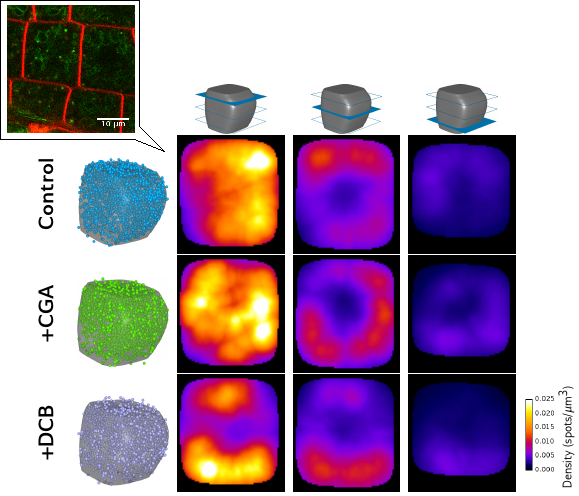 |
Statistical modeling of intra-cellular distributions (Biot et al 2016b) : we developed a pipeline and a software tool to infer, from collections of 3D images, the statistical distribution of intra-cellular compartments within a representative (average) cell. |
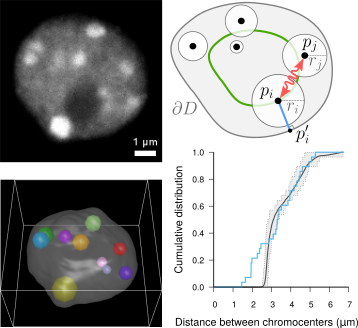 |
Statistical modeling of spatial interactions between biological objects (Andrey et al 2010, Arpon et al 2018, Arpon et al 2021) : we developed a spatial modeling approach to decipher the rules that subtend the spatial distribution of biological objects. Using this approach, we showed that, in Arabidopsis thaliana leaf cell nuclei, chromocenters obey a complex, multiscale organization. |
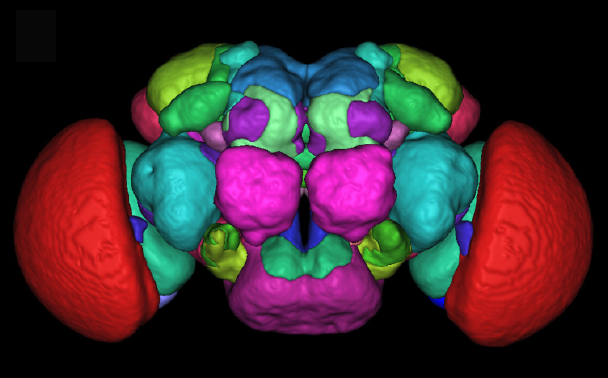 |
A statistical atlas of Drosophila adult brain (Arganda-Carreras et al 2018) : using a collection of annotated images, we built a 3D statistical atlas of Drosophila adult brain and of its main structures. This atlas is a powerful tool to standardize and to compare 3D images of gene expression patterns. |
Leader:
Philippe Andrey