Research teams

Glycans and Signaling
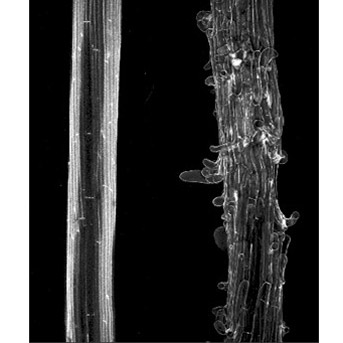
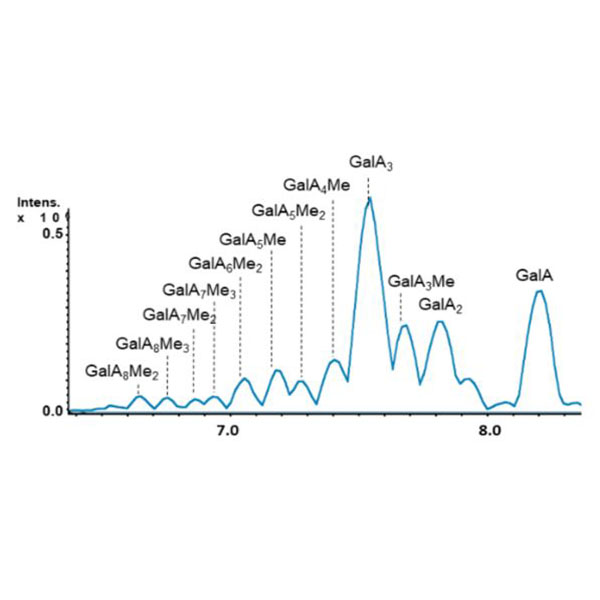
The production of oligosaccharides that comes from the cleavage of cell wall polysaccharides, contributes to the regulation of plant development and growth in physiological and stressed conditions.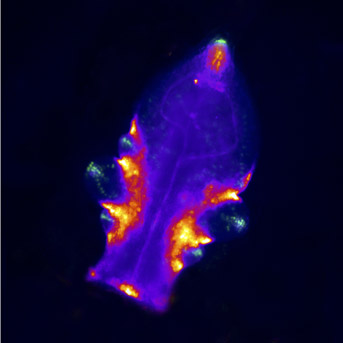
Transcription Factors and Architecture
Our team studies the way plants develop and acquire their remarkable forms. To do this, we are studying the role of transcription factors that orchestrate the development using molecular genetics and imaging approaches combined with quantitative approaches and modeling.
Biology of the Cell and Regeneration
A whole fertile plant can be regenerated starting from a small fragment of tissue, even a single cell. Our studies aim at understanding this process to identify factors that promote or, to the contrary, that impede plant regeneration.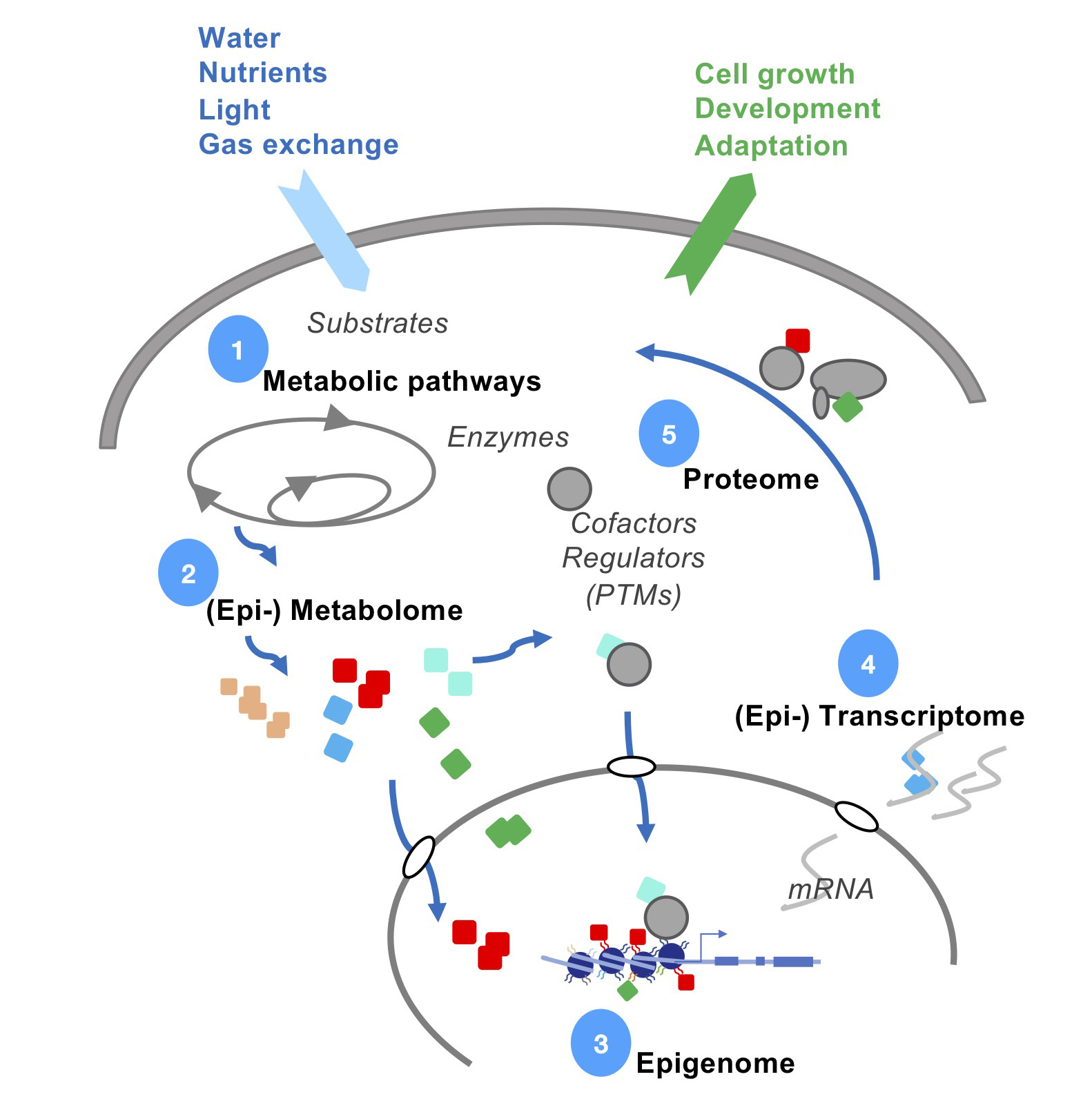
Chromatin Dynamics and Signalling
The CHRODYNO group aims to better understand the inter-process coordination between chromatin dynamics and developmental/environmental signaling by exploiting several biological models and multidisciplinary approaches.
Spatial Control of Cell Division
The SPACE team focuses on the spatial control of cell division, elongation and differentiation in plant cells, in relation to the organization and dynamics of the microtubular cytoskeleton.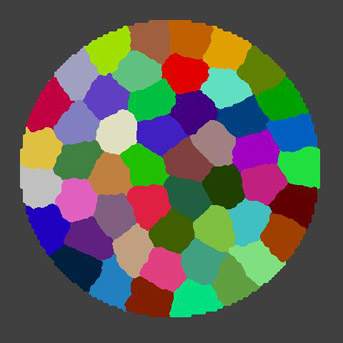
Modeling and Digital Imaging
Our research aims at designing original approaches in image analysis, spatial statistics, and computational modeling to better understand plant development and functions.
Epigenetic Natural Variation
In plants, DNA methylation, that occurs at cytosines in all contexts, is an epigenetic mark critical for transposon silencing. Our group studies the different molecular pathways controlling DNA methylation and its inheritance over generations.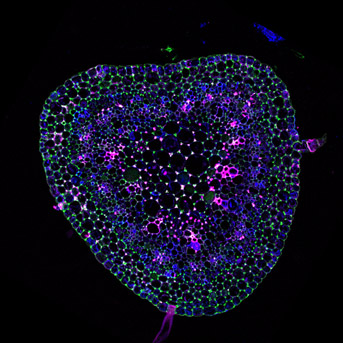
Physiology of plant cell wall assembly, remodelling, and expansion
The team is studying the architecture and metabolism of cell walls in relation to cell expansion, immunity and as a source of natural defense stimulators in plants. It also contributes to the improvement of lignocellulosic biomass crops such as miscanthus.